Module sarra_py.bilan_carbo
View Source
import numpy as np
import xarray as xr
def variable_dict():
"""
Retrieve the dictionary of variables in the dataset with their respective units.
Returns:
dict: A dictionary containing the variables and their units, where the keys are the variable names and the values are the respective units.
"""
variables = {
# climate
"ddj": ["daily thermal time", "°C.j"],
"sdj": ["sum of thermal time since beginning of emergence", "°C.j"],
# phenology
"changePhase": ["indicator of phase transition day", "binary"],
"numPhase": ["number of phenological stage", "arbitrary units"],
"initPhase": ["indicator of performed phase transition", "binary"],
"phasePhotoper": ["photoperiodic phase indicator", "binary"],
"seuilTempPhaseSuivante": ["sum of thermal time needed to reach the next phenological phase", "°C.j"],
"sommeDegresJourPhasePrec": ["sum of thermal time needed to reach the previous phenological phase", "°C.j"],
"seuilTempPhasePrec": ["sum of thermal time needed to reach the previous phenological phase", "°C.j"],
# carbon balance
"assim": ["plant biomass assimilation", "kg/ha"],
"assimPot": ["plant potential biomass assimilation", "kg/ha"],
"bM": ["net growth rate of living biomass", "kg/(m².d)"],
"cM": ["net growth rate of dead biomass", "kg/(m².d)"],
"rdt": ["grain yield", "kg/ha"],
"rdtPot": ["potential grain yield", "kg/ha"],
"reallocation": ["amount of assimilates reallocated to the yield (supply < demand)", "kg/ha"],
"respMaint": ["amount of assimilates consumed by maintainance respiration", "kg/ha"],
"manqueAssim": ["deficit in assimilates (demand - supply)", "kg/ha"],
# biomass
"biomTotStadeFloraison": ["total biomass of the plant at the end of the flowering stage", "kg/ha"],
"biomTotStadeIp": ["total biomass at the panicle initiation stage", "kg/ha"],
"deltaBiomasseAerienne": ["increment of aerial biomass in one day", "kg/(ha.d)"],
"deltaBiomasseFeuilles": ["increment of leaf biomass in one day", "kg/(ha.d)"],
"biomasseAerienne": ["total aerial biomass", "kg/ha"],
"biomasseVegetative": ["total vegetative biomass", "kg/ha"],
"biomasseTotale": ["total biomass", "kg/ha"],
"biomasseTige": ["total stem biomass", "kg/ha"],
"biomasseRacinaire": ["total root biomass", "kg/ha"],
"biomasseFeuille": ["total leaf biomass", "kg/ha"],
"deltaBiomasseTotale": ["increment of total biomass in one day", "kg/(ha.d)"],
# evapotranspiration
"kce": ["fraction of kc attributable to soil evaporation","decimal percentage"],
"kcp": ["fraction of kc attributable to plant transpiration","decimal percentage"],
"kcTot": ["total crop coefficient",""],
"tr": ["actual crop transpiration","mm/d"],
"trPot": ["potential crop transpiration","mm/d"],
"trSurf": ["",""],
# water balance
"consoRur": ["consumption of water stored in the root system", "mm"],
"water_captured_by_mulch" : ["water captured by the mulch in one day","mm"],
"available_water" : ["available water, sum of rainfall and total irrigation for the day","mm"],
"eauTranspi": ["water available for transpiration from the surface reservoir","mm"],
"correctedIrrigation" : ["corrected irrigation amount","mm/d"],
"cstr" : ["drought stress coefficient", "arbitrary unit"],
"dayVrac" : ["modulated daily root growth","mm/day"],
"delta_root_tank_capacity": ["change in root system water reserve","mm"],
"drainage": ["drainage","mm"],
#// "etm": ["evapotranspiration from the soil moisture","mm/d"],
"etp": ["potential evapotranspiration from the soil moisture","mm/d"],
#// "etr": ["reference evapotranspiration","mm/d"],
"evap": ["evaporation from the soil moisture","mm/d"],
"evapPot": ["potential evaporation from the soil moisture","mm/d"],
"FEMcW": ["water fraction in soil volume explored by the root system","none"],
"fesw": ["fraction of available surface water","decimal percentage"],
"irrigTotDay" : ["total irrigation for the day","mm"],
"vRac" : ["reference daily root growth","mm/day"],
"ftsw": ["fraction of transpirable surface water","decimal percentage"],
"runoff" : ["daily water runoff","mm/d"],
"pFact": ["FAO reference for critical FTSW value for transpiration response","none"],
# water tanks
"irrigation_tank_stock" : ["current stock of water in the irrigation tank","mm"], #! renaming stockIrr to irrigation_tank_stock
"mulch_water_stock" : ["water stored in crop residues (mulch)","mm"], #! renaming stockMc to mulch_water_stock
"root_tank_stock": ["current stock of water in the root system tank","mm"], #! renaming stRu to root_tank_stock
"total_tank_capacity": ["total capacity of the root system tank","mm"], #! renaming stRuMax to total_tank_capacity
"stRur": ["",""], # ["previous season's root system tank stock","mm"],
"previous_root_tank_capacity": ["previous season's root system tank capacity","mm"], #! renaming stRurMaxPrec to previous_root_tank_capacity
"previous_root_tank_stock": ["previous day's root system tank stock","mm"],
"stRurSurf": ["surface root system tank stock","mm"],
"surface_tank_stock": ["current stock of water in the surface root system tank","mm"], #! renaming stRuSurf to surface_tank_stock
"stRuSurfPrec": ["previous day's surface root system tank stock","mm"],
"delta_total_tank_stock": ["change in the total root system tank stock","mm"], #! renaming stRuVar to delta_total_tank_stock
"irrigation_tank_capacity" : ["irrigation tank capacity","mm"], #! renaming ruIrr to irrigation_tank_capacity
"ruRac": ["Water column that can potentially be strored in soil volume explored by root system","mm"],
"conv": ["",""],
"KAssim": ["",""],
"dayBiomLeaf": ["daily growth of leaf biomass","kg/ha/d"],
"dRdtPot": ["daily potential demand from yield","kg/ha/d"],
"FeuilleUp": ["",""],
"kRespMaint": ["",""],
"LitFeuille": ["",""],
"nbJourCompte": ["",""],
"nbjStress": ["",""],
"NbUBT": ["",""],
"sla": ["",""],
"stockRac": ["",""],
"sumPP": ["",""],
"TigeUp": ["",""],
"UBTCulture": ["",""],
"lai":["leaf area index","m2/m2"],
# experimental
"Ncrit": ["",""],
}
return variables
def initialize_simulation(data, grid_width, grid_height, duration, paramVariete, paramITK, date_start):
"""
This function initializes variables related to crop growth in the data
xarray dataset. As the rain is the first variable to be initialized in the
data xarray dataset, its dimensions are used to initialize the other
variables.
This code has been adapted from the original InitiationCulture procedure, from the `MilBilanCarbone.pas` code of the
SARRA model.
Args:
data (_type_): _description_ grid_width (_type_): _description_
grid_height (_type_): _description_ duration (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
### variables to be initialized with values from parameters
# from paramVariete : maximum daily thermal time (°C.j) -> #? unused ?
#// data["sommeDegresJourMaximale"] = (data["rain"].dims, np.full(
#// (duration, grid_width, grid_height),
#// (paramVariete["SDJLevee"] + paramVariete["SDJBVP"] + paramVariete["SDJRPR"] + paramVariete["SDJMatu1"] + paramVariete["SDJMatu2"])
#// ))
#// data["sommeDegresJourMaximale"].attrs = {"units":"°C.j", "long_name":"Maximum thermal time"}
# from paramITK : sowing date
data["sowing_date"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), (paramITK["DateSemis"] - date_start).days))
# from paramITK : automatic irrigation indicator
data["irrigAuto"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), paramITK["irrigAuto"]))
data["irrigAuto"].attrs = {"units":"binary", "long_name":"automatic irrigation indicator"}
####### variables qui viennent de initplotMc
# Initial biomass of crop residues (mulch) (kg/ha)
# Biomasse initiale des résidus de culture (mulch) (kg/ha)
# BiomMc := BiomIniMc;
data["biomMc"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), paramITK["biomIniMc"]))
data["biomMc"].attrs = {"units": "kg/ha", "long_name": "Initial biomass of crop residues (mulch)"}
data["biomMc"] = data["biomMc"].astype("float32")
# ?
# StSurf := StockIniSurf;
# data["stSurf"] = np.full((grid_width, grid_height, duration), paramTypeSol["stockIniSurf"])
# ?
# Ltr := 1;
data["ltr"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), 1.0))
data["ltr"] = data["ltr"].astype("float32")
# Initial biomass of stem residues as litter (kg/ha)
# Biomasse initiale des résidus de tiges sous forme de litière (kg/ha)
# LitTiges := BiomIniMc;
data["LitTige"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), paramITK["biomIniMc"]))
data["LitTige"].attrs = {"units": "kg/ha", "long_name": "Initial biomass of stem residues as litter"}
data["LitTige"] = data["LitTige"].astype("float32")
####### fin variables qui viennent de initplotMc
####### variables eau depuis InitPlotMc
# Initializes variables related to crop residues boimass (mulch) in the data
# xarray dataset. This code has been adapted from the original InitPlotMc
# procedure, Bileau.pas code. Comments with tab indentation are from the
# original code. As the rain is the first variable to be initialized in the
# data xarray dataset, its dimensions are used to initialize the other
# variables.
# Soil maximum water storage capacity (mm)
# Capacité maximale de la RU (mm)
# StRurMax := Ru * ProfRacIni / 1000;
#! renaming stRurMax with root_tank_capacity
#// data["stRurMax"] = data["ru"] * paramITK["profRacIni"] / 1000
data["root_tank_capacity"] = (data["rain"].dims, np.repeat(np.array(data["ru"] * paramITK["profRacIni"] / 1000)[np.newaxis,:,:], duration, axis=0))
#// data["stRurMax"].attrs = {"units": "mm", "long_name": "Soil maximum water storage capacity"}
data["root_tank_capacity"].attrs = {"units": "mm", "long_name": "Soil maximum water storage capacity"}
# Maximum water capacity of surface tank (mm)
# Reserve utile de l'horizon de surface (mm)
# RuSurf := EpaisseurSurf / 1000 * Ru;
#! renaming ruSurf with surface_tank_capacity
#// data["ruSurf"] = data["epaisseurSurf"] / 1000 * data["ru"]
data["surface_tank_capacity"] = data["epaisseurSurf"] / 1000 * data["ru"]
#// data["ruSurf"].attrs = {"units": "mm", "long_name": "Maximum water capacity of surface tank"}
data["surface_tank_capacity"].attrs = {"units": "mm", "long_name": "Maximum water capacity of surface tank"}
# ?
# // PfTranspi := EpaisseurSurf * HumPf;
# // StTot := StockIniSurf - PfTranspi/2 + StockIniProf;
# StTot := StockIniSurf + StockIniProf;
# data["stTot"] = np.full((grid_width, grid_height, duration), (paramTypeSol["stockIniSurf"] + paramTypeSol["stockIniProf"]))
#! modifié pour faire correspondre les résultats de simulation, à remettre en place pour un calcul correct dès que possible
# data["stTot"] = np.full((grid_width, grid_height, duration), (paramTypeSol["stockIniProf"]))
#! renaming stTot to total_tank_stock
#// data["stTot"] = data["stockIniProf"]
#//data["total_tank_stock"] = data["stockIniProf"]
#! coorecting total_tank_stock initialization as it did not have the time dimensions that are required as stock evolves through time
data["total_tank_stock"] = (data["rain"].dims, np.repeat(np.array(data["stockIniProf"])[np.newaxis,:,:], duration, axis=0))
#// data["stTot"].attrs = {"units": "mm", "long_name": "?"}
data["total_tank_stock"].attrs = {"units": "mm", "long_name": "?"}
# Soil maximal depth (mm)
# Profondeur maximale de sol (mm)
# ProfRU := EpaisseurSurf + EpaisseurProf;
#! data["profRu"] = data["epaisseurProf"] + data["epaisseurSurf"]
#! data["profRu"].attrs = {"units": "mm", "long_name": "Soil maximal depth"}
# déplacé dans l'initialisation du sol
# Maximum water capacity to humectation front (mm)
# Quantité d'eau maximum jusqu'au front d'humectation (mm)
# // modif 10/06/2015 resilience stock d'eau
# // Front d'humectation egal a RuSurf trop de stress initial
# // Hum := max(StTot, StRurMax);
# Hum := max(RuSurf, StRurMax);
# // Hum mis a profRuSurf
# Hum := max(StTot, Hum);
data["humectation_front"] = (data["rain"].dims, np.full((duration, grid_width, grid_height),
np.maximum(
np.maximum(
#! renaming ruSurf with surface_tank_capacity
#// data["ruSurf"],
data["surface_tank_capacity"].expand_dims({"time":duration}),
#! renaming stRurMax with root_tank_capacity
#// data["stRurMax"],
data["root_tank_capacity"],
),
#! renaming stTot with total_tank_stock
#// data["stTot"],
data["total_tank_stock"],
)
))
data["humectation_front"].attrs = {"units": "mm", "long_name": "Maximum water capacity to humectation front"}
# Previous value for Maximum water capacity to humectation front (mm)
# HumPrec := Hum;
data["previous_humectation_front"] = data["humectation_front"]
# ?
# StRurPrec := 0;
# Previous value for stTot
# StRurMaxPrec := 0;
# //modif 10/06/2015 resilience stock d'eau
#! renaming stTot with total_tank_stock
#! renaminog stRuPrec with previous_total_tank_stock
#// data["stRuPrec"] = data["stTot"]
data["previous_total_tank_stock"] = data["total_tank_stock"]
####### fin variables eau depuis InitPlotMc
# depuis meteo.pas
kpar = 0.5
data["par"] = kpar * data["rg"]
data["par"].attrs = {"units":"MJ/m2", "long_name":"par"}
# crop density
if ~np.isnan(paramVariete["densOpti"]) :
data["rapDensite"] = data["rain"] * 0 + compute_rapDensite(paramITK, paramVariete)
data["rapDensite"].attrs = {"units":"none", "long_name":"sowing density adjustement factor"}
# initialize variables with values at 0
variables = variable_dict()
for variable in variables :
data[variable] = (data["rain"].dims, np.zeros(shape=(duration, grid_width, grid_height)))
data[variable].attrs = {"units":variables[variable][1], "long_name":variables[variable][0]}
data[variable] = data[variable].astype("float32")
return data
def estimate_kcp(j, data, paramVariete):
"""
Estimate the kcp coefficient based on the maximum crop coefficient `kcMax` and plant cover `ltr`.
The computation of `kcp` is based on the EvolKcpKcIni procedure from the biomasse.pas and exmodules 1 & 2.pas files of the original PASCAL code.
Args:
j (int): The starting index for updating `kcp` in the `data` dataset.
data (xarray.Dataset): A dataset containing the data used in the computation of `kcp`. The dataset should contain the following variables:
- 'numPhase': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the number of phases in the crop cycle.
- 'kcp': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the coefficient of crop growth.
- 'ltr': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the plant cover.
paramVariete (dict): A dictionary containing the parameters for estimating `kcp`. The dictionary should contain the following key:
- 'kcMax': A float, representing the maximum crop coefficient.
Returns:
xarray.Dataset: The updated `data` dataset with the new `kcp` values.
"""
data["kcp"][j:,:,:] = np.where(
data["numPhase"][j,:,:] >= 1,
np.maximum(0.3, paramVariete["kcMax"] * (1 - data["ltr"][j,:,:])),
data["kcp"][j,:,:],
)
return data
def estimate_ltr(j, data, paramVariete):
"""
Estimate the fraction of radiation transmitted to the soil `ltr` based on the leaf area index `lai`.
`ltr` is used as a proxy for plant covering of the soil in the water balance calculation, where 1 represents no plant cover and 0 represents full plant cover. `ltr` is computed as an exponential decay function of `lai` with a decay coefficient `kdf`.
This function is adapted from the EvalLtr procedure from the biomasse.pas and exmodules 1 & 2.pas files of the original PASCAL code.
Args:
j (int): The starting index for updating `ltr` in the `data` dataset.
data (xarray.Dataset): A dataset containing the data used in the computation of `ltr`. The dataset should contain the following variables:
- 'lai': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the leaf area index.
- 'ltr': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the fraction of radiation transmitted to the soil.
paramVariete (dict): A dictionary containing the parameters for estimating `ltr`. The dictionary should contain the following key:
- 'kdf': A float, representing the decay coefficient for `ltr`.
Returns:
xarray.Dataset: The updated `data` dataset with the new `ltr` values.
"""
# group 80
data["ltr"][j:,:,:] = np.exp(-paramVariete["kdf"] * data["lai"][j,:,:])
return data
def estimate_KAssim(j, data, paramVariete):
"""
This function calculates the conversion factor `KAssim`, which is used to convert assimilates into biomass.
The value of `KAssim` depends on the phase of the crop.
The conversion factor is calculated based on a lookup table that maps crop phases to values. The crop phase is
determined by the `numPhase` field in the `data` argument, and the corresponding `KAssim` value is set in the
`KAssim` field of the `data` argument.
Args:
j (int): An integer index specifying the time step.
data (xarray.Dataset): A dataset containing the variables used in the calculation of `KAssim`. The dataset
should include the fields `numPhase`, `sdj`, `seuilTemp PhasePrec`, and `seuilTemp PhaseSuivante`. The
`KAssim` field of the dataset will be updated by this function.
paramVariete (dict): A dictionary of parameters. It should include the fields `txAssimBVP`, `txAssimMatu1`,
and `txAssimMatu2`.
Returns:
xarray.Dataset: The updated `data` dataset, with the `KAssim` field set to the calculated values.
"""
phase_equivalences = {
2: 1,
3: paramVariete['txAssimBVP'],
4: paramVariete['txAssimBVP'],
#! replacing sommeDegresJourPhasePrec with seuilTempPhasePrec
#// 5: paramVariete["txAssimBVP"] + (data['sdj'][j,:,:] - data['sommeDegresJourPhasePrec'][j,:,:]) * (paramVariete['txAssimMatu1'] - paramVariete['txAssimBVP']) / (data['seuilTempPhaseSuivante'][j,:,:] - data['sommeDegresJourPhasePrec'][j,:,:]),
5: paramVariete["txAssimBVP"] + (data['sdj'][j,:,:] - data['seuilTempPhasePrec'][j,:,:]) * (paramVariete['txAssimMatu1'] - paramVariete['txAssimBVP']) / (data['seuilTempPhaseSuivante'][j,:,:] - data['seuilTempPhasePrec'][j,:,:]),
#// 6: paramVariete["txAssimMatu1"] + (data["sdj"][j,:,:] - data["sommeDegresJourPhasePrec"][j,:,:]) * (paramVariete["txAssimMatu2"] - paramVariete["txAssimMatu1"]) / (data["seuilTempPhaseSuivante"][j,:,:] - data["sommeDegresJourPhasePrec"][j,:,:]),
6: paramVariete["txAssimMatu1"] + (data["sdj"][j,:,:] - data["seuilTempPhasePrec"][j,:,:]) * (paramVariete["txAssimMatu2"] - paramVariete["txAssimMatu1"]) / (data["seuilTempPhaseSuivante"][j,:,:] - data["seuilTempPhasePrec"][j,:,:]),
}
for phase in range(2,7):
data["KAssim"][j:,:,:] = np.where(
data["numPhase"][j,:,:] == phase,
phase_equivalences[phase],
data["KAssim"][j,:,:],
)
return data
def estimate_conv(j,data,paramVariete):
"""
This function calculates the conversion of assimilates into biomass.
The conversion factor is determined by multiplying the KAssim value, which
is dependent on the phase of the crop, with the conversion rate (txConversion)
specified in the `paramVariete` argument.
Args:
j (int): The starting index of the calculation
data (dict): A dictionary containing information on the crop growth, including
the phase of the crop and the KAssim value.
paramVariete (dict): A dictionary containing parameters relevant to the crop
growth, including the conversion rate.
Returns:
dict: The input `data` dictionary with the calculated "conv" value added.
"""
data["conv"][j:,:,:] = (data["KAssim"][j,:,:] * paramVariete["txConversion"])
return data
def BiomDensOptSarraV4(j, data, paramITK):
"""
si densité plus faible alors on considére qu'il faut augmenter les biomasses, LAI etc
en regard de cette situation au niveau de chaque plante (car tout est rapporté é des kg/ha).
Si elle est plus forte on ne change rien pour lors.
Valeur fixe en ref au maés é déf en paramétre par variétésé rapDensite := Max(1, 70000/densite);
"""
"""
if ~np.isnan(paramVariete["densOpti"]) :
paramITK["rapDensite"] = np.maximum(1,paramVariete["densOpti"]/paramITK["densite"])
data["rdt"][j,:,:] = data["rdt"][j,:,:] * paramITK["rapDensite"]
data["biomasseRacinaire"][j,:,:] = data["biomasseRacinaire"][j,:,:] * paramITK["rapDensite"]
data["biomasseTige"][j,:,:] = data["biomasseTige"][j,:,:] * paramITK["rapDensite"]
data["biomasseFeuille"][j,:,:] = data["biomasseFeuille"][j,:,:] * paramITK["rapDensite"]
data["biomasseAerienne"][j,:,:] = data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:]
data["lai"][j,:,:] = data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:]
data["biomasseTotale"][j,:,:] = data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:]
return data
"""
return data
def compute_rapDensite(paramITK, paramVariete):
"""
It basically calculates a correction factor (rapDensite).
This correction factor Is calculated with an equation of form
a + p * exp( -(x/(o / -log((1-a)/p) )) )
with a the densiteA parameter
p the densiteP parameter$
x the actual crop density
o the densOpti parameter
See
https://www.wolframalpha.com/input?i=a+%2B+p+*+exp%28-%28x+%2F+%28+o%2F-+log%28%281+-+a%29%2F+p%29%29%29%29
for equation visualization.
This equation is probably too complex for the problem at hand.
Args:
j (_type_): _description_
data (_type_): _description_
paramITK (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
rapDensite = paramVariete["densiteA"] + paramVariete["densiteP"] * np.exp(-(paramITK["densite"] / ( paramVariete["densOpti"]/- np.log((1 - paramVariete['densiteA'])/ paramVariete["densiteP"]))))
return rapDensite
def adjust_for_sowing_density(j, data, paramVariete, direction):
"""
This function translates the effect of sowing density on biomass and LAI.
This function is adapted from the BiomDensOptSarV42 and BiomDensiteSarraV42
procedures, from the bilancarbonsarra.pas original Pascal code.
Notes from CB :
if density is lower than the optimal density, then we consider that we need
to increase the biomass, LAI etc in regard of this situation at each plant
level (because everything is related to kg/ha). If it is higher, it
increases asymptotically.
Args:
j (_type_): _description_
data (_type_): _description_
paramITK (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
if direction == "in" :
if ~np.isnan(paramVariete["densOpti"]) :
data["rdt"][j:,:,:] = data["rdt"][j,:,:] * data["rapDensite"][j,:,:]
data["rdtPot"][j:,:,:] = (data["rdtPot"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseRacinaire"][j:,:,:] = (data["biomasseRacinaire"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseTige"][j:,:,:] = (data["biomasseTige"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseFeuille"][j:,:,:] = (data["biomasseFeuille"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseAerienne"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:])
data["lai"][j:,:,:] = (data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:])
data["biomasseTotale"][j:,:,:] = (data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:])
return data
if direction == "out":
if ~np.isnan(paramVariete["densOpti"]):
data["rdt"][j:,:,:] = (data["rdt"][j,:,:] / data["rapDensite"][j,:,:])
data["rdtPot"][j:,:,:] = (data["rdtPot"][j,:,:]/ data["rapDensite"][j,:,:])
data["biomasseRacinaire"][j:,:,:] = (data["biomasseRacinaire"][j,:,:] / data["rapDensite"][j,:,:])
data["biomasseTige"][j:,:,:] = (data["biomasseTige"][j,:,:] / data["rapDensite"][j,:,:])
data["biomasseFeuille"][j:,:,:] = (data["biomasseFeuille"][j,:,:] / data["rapDensite"][j,:,:])
data["biomasseAerienne"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:])
#? conflit avec fonction evolLAIphase ?
#data["lai"][j:,:,:] = data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:]
data["lai"][j:,:,:] = data["lai"][j:,:,:] / data["rapDensite"][j,:,:]
data["biomasseTotale"][j:,:,:] = (data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:])#[...,np.newaxis]
#data["biomasseTotale"][j:,:,:] = data["biomasseTotale"][j:,:,:] / data["rapDensite"]
return data
def EvalAssimSarrahV4(j, data):
"""
data["parIntercepte"][j,:,:] = 0.5 * (1 - data["ltr"][j,:,:]) * data["rg"][j,:,:]
data["assimPot"][j:,:,:] = data["parIntercepte"][j,:,:] * data["conv"][j,:,:] * 10
data["assim"][j,:,:] = np.where(
data["trPot"][j,:,:] > 0,
data["assimPot"][j,:,:] * data["tr"][j,:,:] / data["trPot"][j,:,:],
0,
)
"""
return data
def update_assimPot(j, data, paramVariete, paramITK):
"""
Update the assimPot value based on the intensification level (NI).
If the intensification level `NI` is defined in `paramITK`, the conversion rate `txConversion` is computed using a formula based on `NIYo`, `NIp`, `LGauss`, and `AGauss`. If `NI` is not defined, `assimPot` is updated using `conv`, which is updated in the `estimate_conv` function using the variables `KAssim` and `txConversion`.
When NI parameter is used (from to 4), conversion rate txConversion
is computed using the following formula :
NIYo + NIp * (1-exp(-NIp * NI)) - (exp(-0.5*((NI - LGauss)/AGauss)* (NI- LGauss)/AGauss))/(AGauss*2.506628274631)
This function is adapted from the `EvalAssimSarraV42` procedure in the `bilancarbonsarra.pas` file of the original Pascal code.
Note from
CB : correction of the conversion rate depending on the intensification
level
notes from CB reharding the EvalAssimSarraV42 procedure :
Modif du 04/03/2021 : Prise en compte en plus de la densit� de semis de
l'effet niveau d'intensification NI NI = 1 quand on est � l'optimum du
niveau d'intensification. Dans le cas de situation contr�l� c'est la
fertilit� qui est la clef principale en prenant en r�f�rence la qt� d'azote
(�quivalent phosphore...) optimum Il peut aller � 0 ou �tre sup�rieur � 1 si
situation sur optimum, ie un peu plus de rdt mais � cout trop �lev�... On
�value un nouveau tx de conversion en fn du Ni au travers d'une double
�quation : asympote x gaussienne invers�e Et d'un NI d�fini en fn du
sc�nario de simulation ou des donn�es observ�es. NIYo = D�calage en Y de
l'asymptote NIp = pente de l'asymptote LGauss = Largeur de la Guaussienne
AGauss = Amplitude de la Guaussienne
Conversion qui est la valeur du taux de conversion en situation optimum n'a
plus besoin d'�tre utilis� sinon dans la calibration des param�tres de cette
�quation en absence de donn�es sur ces param�tres on ne met aucune valeur �
NI CF fichier ex IndIntensite_txConv_eq.xls}
Args:
- j (int): An index that represents the current iteration.
- data (dict): A dictionary containing data arrays with the following keys:
- "assimPot" (np.ndarray): An array representing the potential assimilation rate.
- "par" (np.ndarray): An array representing photosynthetically active radiation.
- "lai" (np.ndarray): An array representing the leaf area index.
- "conv" (np.ndarray): An array representing the conversion rate.
- paramVariete (dict): A dictionary containing parameters for the computation of the conversion rate, including:
- "txConversion" (float): The conversion rate.
- "NIYo" (float): The shift in the Y-axis of the asymptote.
- "NIp" (float): The slope of the asymptote.
- "LGauss" (float): The width of the Gaussian curve.
- "AGauss" (float): The amplitude of the Gaussian curve.
- "kdf" (float): The constant used in the computation of `assimPot`.
- paramITK (dict): A dictionary containing the intensification level `NI` (float).
Returns:
- data (dict): The input `data` dictionary with the updated "assimPot" array.
"""
if ~np.isnan(paramITK["NI"]):
#? the following (stupidly long) line was found commented, need to check why and if this is correct
paramVariete["txConversion"] = paramVariete["NIYo"] + paramVariete["NIp"] * (1-np.exp(-paramVariete["NIp"] * paramITK["NI"])) - (np.exp(-0.5*((paramITK["NI"] - paramVariete["LGauss"])/paramVariete["AGauss"])* (paramITK["NI"]- paramVariete["LGauss"])/paramVariete["AGauss"]))/(paramVariete["AGauss"]*2.506628274631)
# NIYo + NIp * (1-exp(-NIp * NI)) - (exp(-0.5*((NI - LGauss)/AGauss)* (NI- LGauss)/AGauss))/(AGauss*2.506628274631)
data["assimPot"][j,:,:] = data["par"][j,:,:] * \
(1-np.exp(-paramVariete["kdf"] * data["lai"][j,:,:])) * \
paramVariete["txConversion"] * 10
else :
data["assimPot"][j,:,:] = data["par"][j,:,:] * \
(1-np.exp(-paramVariete["kdf"] * data["lai"][j,:,:])) * \
data["conv"][j,:,:] * 10
return data
def update_assim(j, data):
"""
This function updates assim. If trPot (potential transpiration from the
plant, mm) is greater than 0, then assim equals assimPot, multiplied by the
ratio of effective transpiration over potential transpiration.
If potential transpiration is null, then assim is null as well.
Is it adapted from the EvalAssimSarraV42 procedure, of the
bilancarbonsarra.pas file from the original Pascal code
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["assim"][j,:,:] = np.where(
data["trPot"][j,:,:] > 0,
data["assimPot"][j,:,:] * data["tr"][j,:,:] / data["trPot"][j,:,:],
0,
)
return data
def calculate_maintainance_respiration(j, data, paramVariete):
"""
This function calculates the maintenance respiration `respMaint` (in kg/ha/j in equivalent dry matter) of the plant.
The maintenance respiration is calculated by summing the maintenance respiration associated with total biomass and
leaves biomass. If the plant's growth phase is above 4 and there is no leaf biomass, `respMaint` is set to 0.
The calculation is based on the equation:
coefficient_temp = 2^((average_temp - tempMaint) / 10)
respiration = kRespMaint * biomass * coefficient_temp
where `average_temp` is the average temperature for the day, `tempMaint` is the maintenance temperature from
`variety_params`, `kRespMaint` is the maintenance respiration coefficient from `variety_params`, and `biomass`
is the total or leaf biomass.
Args:
j (int): The time step of the calculation.
data (xarray.Dataset): The input data containing the variables `tpMoy`, `biomasseTotale`, `biomasseFeuille`, and
`numPhase`. The output `respMaint` will also be stored in this dataset.
variety_params (dict): The parameters related to the plant variety, containing the keys `tempMaint` and
`kRespMaint`.
Returns:
xarray.Dataset: The input `data` with the updated `respMaint` variable.
"""
coefficient_temp = 2**((data["tpMoy"][j,:,:] - paramVariete["tempMaint"]) / 10)
resp_totale = paramVariete["kRespMaint"] * data["biomasseTotale"][j,:,:] * coefficient_temp
resp_feuille = paramVariete["kRespMaint"] * data["biomasseFeuille"][j,:,:] * coefficient_temp
data["respMaint"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 4) & (data["biomasseFeuille"][j,:,:]==0),
0,
resp_totale + resp_feuille,
)
return data
def update_total_biomass(j, data, paramVariete, paramITK):
"""
Update the Total Biomass of the Plant.
The total biomass is updated based on the plant's current phase and other parameters.
If the plant is in phase 2 and there's a change in phase, the total biomass is initialized using
crop density, grain yield per plant, and the dry weight of the grain.
If the plant is not in phase 2 or there's no change in phase, the total biomass is incremented with
the difference between the plant's assimilation and maintenance respiration.
When passing from phase 1 to phase 2, total biomass is initialized.
Initialization value is computed from crop density (plants/ha), txResGrain
(grain yield per plant), and poidsSecGrain. Otherwise, total biomass is
incremented with the difference between plant assimilation assim and
maintainance respiration respMaint.
This function is adapted from the EvolBiomTotSarrahV4 procedure, of the
bilancarbonsarra.pas file from the original Pascal code.
Args:
j (int): The current time step.
data (xarray.Dataset): The data for the plant, including variables like
"biomasseTotale", "assim", "respMaint", "numPhase", and "changePhase".
paramVariete (dict): A dictionary of parameters specific to the plant variety.
paramITK (dict): A dictionary of inter-tropical convergence zone parameters.
Returns:
xarray.Dataset: The updated data for the plant, including the updated "biomasseTotale"
and "deltaBiomasseTotale" variables.
"""
data["biomasseTotale"][j:,:,:] = np.where(
(data["numPhase"][j,:,:]==2) & (data["changePhase"][j,:,:]==1),
paramITK["densite"] * np.maximum(1,paramVariete['densOpti']/paramITK['densite']) * paramVariete["txResGrain"] * paramVariete["poidsSecGrain"] / 1000,
data["biomasseTotale"][j,:,:] + (data["assim"][j,:,:] - data["respMaint"][j,:,:]),
)
# we may want to drop this variable and use the raw computation instead
data["deltaBiomasseTotale"][j:,:,:] = (data["assim"][j,:,:] - data["respMaint"][j,:,:])
return data
def update_total_biomass_stade_ip(j, data):
"""
Update the total biomass of the plant at the end of the vegetative phase (ip = "initiation paniculaire").
If the plant has reached phase 4 and has just changed phase, the current
total biomass will be copied to the "biomTotStadeIp" variable, which represents
the total biomass at the end of the vegetative phase (initiation paniculaire).
This function is adapted from the EvalRdtPotRespSarV42 procedure, of
the bilancarbonsarra.pas file from the original Pascal code.
Args:
j (int): Timestep index.
data (xarray.Dataset): Input dataset.
Returns:
xarray.Dataset: The updated dataset with the "biomTotStadeIp" variable updated.
"""
data["biomTotStadeIp"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 4) & (data["changePhase"][j,:,:] == 1),
data["biomasseTotale"][j,:,:],
data["biomTotStadeIp"][j,:,:],
)
return data
def update_total_biomass_at_flowering_stage(j, data):
"""
This function updates the total biomass of the plant at the end of the
flowering stage (biomTotStadeFloraison).
If the plant is in phase 5, and the phase has changed, then the total
biomass is copied to the biomTotStadeFloraison variable.
This function is adapted from the EvalRdtPotRespSarV42 procedure, of
the bilancarbonsarra.pas file from the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomTotStadeFloraison"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5) & (data["changePhase"][j,:,:] == 1),
data["biomasseTotale"][j,:,:],
data["biomTotStadeFloraison"][j,:,:],
)
return data
def update_potential_yield(j, data, paramVariete):
"""
Update the potential yield of the plant.
The potential yield is initialized as an affine function of the delta
between the end of the vegetative phase and the end of the flowering stage,
plus a linear function of the total biomass at the end of the flowering stage.
The potential yield is capped to twice the biomass of the stem to avoid unrealistic
values.
The update occurs if the plant is in phase 5 and its phase has changed
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the
bilancarbonsarra.pas file from the original Pascal code.
Args:
j (int): An index representing the current time step.
data (xarray.Dataset): A dataset containing plant data.
paramVariete (dict): A dictionary containing parameters for the plant variety.
Returns:
xarray.Dataset: The input `data` with the potential yield updated.
"""
delta_biomass_flowering_ip = data["biomTotStadeFloraison"][j,:,:] - data["biomTotStadeIp"][j,:,:]
data["rdtPot"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5) & (data["changePhase"][j,:,:] == 1),
(paramVariete["KRdtPotA"] * delta_biomass_flowering_ip + paramVariete["KRdtPotB"]) + paramVariete["KRdtBiom"] * data["biomTotStadeFloraison"][j,:,:],
data["rdtPot"][j,:,:],
)
#! phaseDevVeg pas utilisé ? attention c'est un paramètre variétal et pas un jeu de donées
data["rdtPot"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5) & (data["changePhase"][j,:,:] == 1) & (data["rdtPot"][j,:,:] > data["biomasseTige"][j,:,:] * 2) & (paramVariete["phaseDevVeg"] < 6),
data["biomasseTige"][j,:,:] * 2,
data["rdtPot"][j,:,:],
)
return data
def update_potential_yield_delta(j, data, paramVariete):
"""
This function updates the delta potential yield (dRdtPot) of the plant, which is the rate at which the
plant's yield is changing over time. The delta potential yield is calculated as the product of the potential
yield, the ratio of actual degree days to maturity, and the ratio of actual transpiration to potential transpiration.
The calculation is only done if the plant is in phase 5 and the potential transpiration is above 0.
If the potential transpiration is not above 0, the delta potential yield is set to 0.
For all other phases, the delta potential yield is unchanged.
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the
bilancarbonsarra.pas file from the original Pascal code.
Args:
- j (int): an integer index, representing the current step of the simulation
- data (xarray dataset): the simulation data, including the current state of the plant
- paramVariete (dict): the variety-specific parameters used in the simulation
Returns:
- data (xarray dataset): the updated simulation data, including the updated delta potential yield
"""
data["dRdtPot"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5),
np.where(
(data["trPot"][j,:,:] > 0),
np.maximum(
data["rdtPot"][j,:,:] * (data["ddj"][j,:,:] / paramVariete["SDJMatu1"]) * (data["tr"][j,:,:] / data["trPot"][j,:,:]),
data["respMaint"][j,:,:] * 0.15,
),
0,
),
data["dRdtPot"][j,:,:],
)
return data
def update_aboveground_biomass(j, data, paramVariete):
"""
Update the aboveground biomass of the plant.
The aboveground biomass is either updated based on a linear function of the total biomass, if the plant is in phase 2, 3, or 4, or incremented with the total biomass delta if the plant is in any other phase.
This function is based on the EvolBiomAeroSarrahV3 procedure, of the
***bilancarbonsarra***, exmodules 1 & 2.pas file from the original Pascal
code.
Args:
j (int): The current iteration step in the simulation.
data (xarray.Dataset): The simulation data, including the current phase of the plant and various biomass values.
paramVariete (dict): The parameters of the plant variety.
Returns:
xarray.Dataset: The updated simulation data, including the updated aboveground biomass and delta aboveground biomass.
"""
#// data["deltaBiomasseAerienne"][j:,:,:] = np.copy(data["biomasseAerienne"][j,:,:])
data["biomasseAerienne"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] >= 2) & (data["numPhase"][j,:,:] <= 4),
np.minimum(0.9, paramVariete["aeroTotPente"] * data["biomasseTotale"][j,:,:] + paramVariete["aeroTotBase"]) * data["biomasseTotale"][j,:,:],
data["biomasseAerienne"][j,:,:] + data["deltaBiomasseTotale"][j,:,:],
)
#//data["deltaBiomasseAerienne"][j:,:,:] = (data["biomasseAerienne"][j,:,:] - data["deltaBiomasseAerienne"][j,:,:])#[...,np.newaxis]
data["deltaBiomasseAerienne"][j:,:,:] = data["biomasseAerienne"][j,:,:] - data["biomasseAerienne"][j-1,:,:]
return data
def estimate_reallocation(j, data, paramVariete):
"""
Estimate the daily biomass reallocation between stem and leaves.
This function computes the daily biomass reallocation between stem and leaves for the plant. The computation
only occurs when the plant is in phase 5. The amount of biomass that can be reallocated is estimated as
follows:
1. The difference between the potential yield delta and the aboveground biomass delta, bound by 0, is
calculated and referred to as manqueAssim. manqueAssim represents the daily variation in biomass that
remains after the plant has built its aboveground biomass.
2. The reallocation is computed as the minimum of the product of manqueAssim and the reallocation rate and
the difference between the leaf biomass and 30, also bound by 0. The value of 30 is an arbitrary
threshold which ensures that reallocation is 0 if the leaf biomass is below 30. If the leaf biomass is
above 30, reallocation is bounded by biomasseFeuille - 30.
If the plant is not in phase 5, reallocation is set to 0.
This function is based on the EvalReallocationSarrahV3 procedure from the bilancarbonsarra.pas and
exmodules 1 & 2.pas files from the original Pascal code.
Args:
j (int): Current time step of the simulation.
data (xarray.Dataset): The dataset containing all the simulation data.
paramVariete (dict): A dictionary containing the parameters for the simulation.
Returns:
xarray.Dataset: The updated dataset with the reallocation values.
"""
condition = (data["numPhase"][j,:,:] == 5)
data["manqueAssim"][j:,:,:] = np.where(
condition,
np.maximum(0, (data["dRdtPot"][j,:,:] - np.maximum(0.0, data["deltaBiomasseAerienne"][j,:,:]))),
0,
)
data["reallocation"][j:,:,:] = np.where(
condition,
np.minimum(
data["manqueAssim"][j,:,:] * paramVariete["txRealloc"],
np.maximum(0.0, data["biomasseFeuille"][j,:,:] - 30),
),
0,
)
return data
def update_root_biomass(j, data):
"""
Update the root biomass (biomasseRacinaire) for a given time step.
The root biomass is computed as the difference between the total biomass
and the aboveground biomass.
This function is based on the EvalBiomasseRacinaire procedure, of the
milbilancarbone, exmodules 1 & 2, ***milbilancarbone***.pas file from the
original Pascal code
Args:
j (int): Time step index.
data (xarray.Dataset): Input dataset containing relevant variables.
Returns:
xarray.Dataset: Updated dataset with the root biomass variable.
"""
data["biomasseRacinaire"][j,:,:] = data["biomasseTotale"][j,:,:] - data["biomasseAerienne"][j,:,:]
return data
def update_leaf_biomass(j, data, paramVariete):
"""
For phase above 1 and if the delta of aerial biomass is negative,
meaning that the plant is losing aerial biomass, the leaf biomass is
updated as the difference between the leaf biomass and the reallocation
minus the delta of aerial biomass multiplied by the reallocation rate in
leaves. This value is bound in 0.00000001.
Otherwise, the leaf biomass is not updated.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of
the bilancarbonsarra.pas and exmodules 1 & 2.pas files from the original
Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseFeuille"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] < 0),
np.maximum(
0.00000001,
data["biomasseFeuille"][j,:,:] - (data["reallocation"][j,:,:] - data["deltaBiomasseAerienne"][j,:,:]) * paramVariete["pcReallocFeuille"]
),
data["biomasseFeuille"][j,:,:],
)
return data
def update_stem_biomass(j, data, paramVariete):
"""
For phase above 1 and if the delta of aerial biomass is negative,
meaning that the plant is losing aerial biomass, the stem biomass is
updated as the difference between the leaf biomass and the reallocation
minus the delta of aerial biomass multiplied by (1-reallocation rate in
leaves) (if it's not leaves, it's stems...). This value is bound in 0.00000001.
Otherwise, the stem biomass is not updated.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of
the bilancarbonsarra.pas and exmodules 1 & 2.pas files from the original
Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
# group 122
data["biomasseTige"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] < 0),
np.maximum(
0.00000001,
data["biomasseTige"][j,:,:] - (data["reallocation"][j,:,:] - data["deltaBiomasseAerienne"][j,:,:]) * (1 - paramVariete["pcReallocFeuille"]),
),
data["biomasseTige"][j,:,:],
)
return data
def condition_positive_delta_biomass(j, data, paramVariete):
condition = (data["numPhase"][j,:,:] > 1) & \
(data["deltaBiomasseAerienne"][j,:,:] >= 0) & \
((data["numPhase"][j,:,:] <= 4) | (data["numPhase"][j,:,:] <= paramVariete["phaseDevVeg"]))
# (data["numPhase"][j,:,:] <= 4)
return condition
def update_bM_and_cM(j, data, paramVariete):
"""
This function returns the updated values of bM and cM.
bM and cM are updated if the delta of aerial biomass is positive,
meaning that the plant is gaining aerial biomass, and if the phase is
above 1 and below 4 or the phase is below the vegetative phase.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of
the bilancarbonsarra.pas files from the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["bM"][j,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
paramVariete["feuilAeroBase"] - 0.1,
data["bM"][j,:,:],
)
data["cM"][j,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
((paramVariete["feuilAeroPente"] * 1000)/ data["bM"][j,:,:] + 0.78) / 0.75,
data["cM"][j,:,:],
)
return data
def update_leaf_biomass_positive_delta_aboveground_biomass(j, data, paramVariete):
"""
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseFeuille"][j:,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
(0.1 + data["bM"][j,:,:] * data["cM"][j,:,:] ** ((data["biomasseAerienne"][j,:,:] - data["rdt"][j,:,:]) / 1000)) \
* (data["biomasseAerienne"][j,:,:] - data["rdt"][j,:,:]),
data["biomasseFeuille"][j,:,:],
)
return data
def update_stem_biomass_positive_delta_aboveground_biomass(j, data, paramVariete):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseTige"][j:,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
data["biomasseAerienne"][j,:,:] - data["biomasseFeuille"][j,:,:] - data["rdt"][j,:,:],
data["biomasseTige"][j,:,:],
)
return data
def condition_positive_delta_aboveground_biomass_all_phases(j, data):
#// condition = (data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] >= 0)
condition = (data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] > 0)
return condition
def update_leaf_biomass_all_phases(j, data, paramVariete):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseFeuille"][j:,:,:] = np.where(
condition_positive_delta_aboveground_biomass_all_phases(j, data),
data["biomasseFeuille"][j,:,:] - data["reallocation"][j,:,:] * paramVariete["pcReallocFeuille"],
data["biomasseFeuille"][j,:,:],
)
return data
def update_stem_biomass_all_phases(j, data, paramVariete):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseTige"][j:,:,:] = np.where(
condition_positive_delta_aboveground_biomass_all_phases(j, data),
data["biomasseTige"][j,:,:] - (data["reallocation"][j,:,:] * (1- paramVariete["pcReallocFeuille"])),
data["biomasseTige"][j,:,:],
)
return data
def update_aboveground_biomass_step_2(j, data):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseAerienne"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 1),
data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:],
data["biomasseAerienne"][j,:,:],
)
return data
def EvalFeuilleTigeSarrahV4(j, data, paramVariete):
"""
This function is a wrapper
It is adapted from the EvalFeuilleTigeSarrahV4 procedure from the bilancarbonsarra.pas file
of the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
# data["deltaBiomasseFeuilles"][j:,:,:] = np.where(
# (data["numPhase"][j,:,:] > 1),
# data["biomasseFeuille"][j,:,:],
# data["deltaBiomasseFeuilles"][j,:,:],
# )
# if (data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] < 0)
data = update_leaf_biomass(j, data, paramVariete)
data = update_stem_biomass(j, data, paramVariete)
# if deltaBiomasseAerienne >= 0 and (numPhase <= 4 or numPhase <= phaseDevVeg)
data = update_bM_and_cM(j, data, paramVariete)
data = update_leaf_biomass_positive_delta_aboveground_biomass(j, data, paramVariete)
data = update_stem_biomass_positive_delta_aboveground_biomass(j, data, paramVariete)
# if deltaBiomasseAerienne > 0 and numPhase > 1
data = update_leaf_biomass_all_phases(j, data, paramVariete)
data = update_stem_biomass_all_phases(j, data, paramVariete)
# condition = (data["numPhase"][j,:,:] > 1)
# data["deltaBiomasseFeuilles"][j:,:,:] = np.where(
# (data["numPhase"][j,:,:] > 1),
# data["biomasseFeuille"][j,:,:] - data["deltaBiomasseFeuilles"][j,:,:],
# data["deltaBiomasseFeuilles"][j,:,:],
# )
# simpler formulation for updating the deltaBiomasseFeuilles
data["deltaBiomasseFeuilles"][j:,:,:] = data["biomasseFeuille"][j,:,:] - data["biomasseFeuille"][j-1,:,:]
data = update_aboveground_biomass_step_2(j, data)
return data
def update_vegetative_biomass(j, data):
"""_summary_
This function is adapted from the EvalBiomasseVegetati procedure from the copie milbilancarbon, exmodules 1 & 2, ***milbilancarbone*** file
of the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseVegetative"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:])
return data
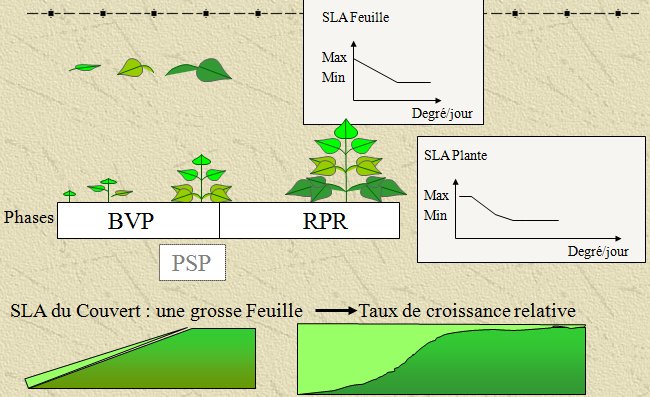
def calculate_canopy_specific_leaf_area(j, data, paramVariete):
"""
Calculate the specific leaf area (SLA) of the canopy.
If the leaf biomass is positive, and if we are at the transition day between
phases 1 and 2 (numPhase = 2 and changePhase = 1), then the SLA is set to
`slaMax`.
If the leaf biomass is positive and increasing (deltaBiomasseFeuilles is
positive), the SLA for existing leaves is calculated by reducing it by an
amount proportional to the current SLA, while the SLA for new leaves is
calculated as the average between SLA and `slaMax`. The SLA for the entire
canopy is then calculated as the weighted average of the SLAs for existing
and new leaves.
If there is no increase in leaf biomass (deltaBiomasseFeuilles is negative),
only the SLA for existing leaves is calculated.
If the leaf biomass is negative, the SLA is unchanged.
The calculated SLA value is bounded between `slaMin` and `slaMax`.
This function is adapted from the EvalSlaSarrahV3 procedure in the
bilancarbonsarra.pas and exmodules 1 & 2.pas files of the original Pascal
code. This calculation method assumes that young leaves have a higher SLA
than old leaves and that the fraction of young leaves makes the canopy SLA
increase. The `penteSLA` parameter causes a general decrease in SLA
(penteSLA = relative decrease per day = fraction of difference between
`slaMax` and `slaMin`).
Expected parameters:
SLAmax [0.001, 0.01]
SLAmin [0.001, 0.01]
penteSLA [0, 0.2]
SLAini = SLAmax
This function estimates the specific leaf area (SLA) of the canopy.
First, if the leaf biomass is positive, if numPhase = 2 and changePhase = 1,
which means we are at the transition day between phases 1 and 2, sla is set
to be equal to slaMax.
Then, if the leaf biomass is positive, and if deltaBiomasseFeuilles is
positive (meaning that the leaf biomass is increasing), SLA for already
existing leaves is calculated by removing a value that is an affine function
of SLA itself, and SLA for new leaves is calculated as the mean between SLA
and slaMax ; then the SLA is calculated as the weighted mean of the two SLA
values.
Logically, if there is no newly produced leaf biomass (deltaBiomasseFeuilles
is negative), only the SLA for already existing leaves is calculated.
If biomasseFeuille is negative, SLA is unchanged.
Finally, if biomasseFeuille is positive, SLA value is bounded between slaMin
and slaMax.
This function is adapted from the EvalSlaSarrahV3 procedure from the
bilancarbonsarra.pas and exmodules 1 & 2.pas file of the original Pascal
code. We note that multiple versions of the calculation methods have been
used in the original procecure. We may want to go back to that if this
function is problematic.
Notes :
In this approach, it is assumed that young leaves have a higher SLA than old
leaves. The fraction of young leaves makes the canopy SLA increase. The
penteSLA parameter causes a general decrease in SLA (penteSLA = relative
decrease per day = fraction of difference between SLAmax and SLAmin). This
approach is known for legumes, but can also be adapted to other species.
Generic/expected parameters :
SLAmax [0.001, 0.01]
SLAmin [0.001, 0.01]
penteSLA [0, 0.2]
SLAini = SLAmax
Args:
- j (int): The time step.
- data (xarray.Dataset): The data for all variables.
- paramVariete (dict): Parameters for the calculation.
Returns:
- data (xarray.Dataset): The updated data with the calculated SLA.
"""
condition = (data["biomasseFeuille"][j,:,:] > 0) & \
(data["numPhase"][j,:,:] == 2) & \
(data["changePhase"][j,:,:] == 1)
data["sla"][j:,:,:] = np.where(
condition,
paramVariete["slaMax"],
data["sla"][j,:,:],
)
ratio_old_leaf_biomass = data["biomasseFeuille"][j-1,:,:] / data["biomasseFeuille"][j,:,:]
ratio_new_leaf_biomass = data["deltaBiomasseFeuilles"][j,:,:] / data["biomasseFeuille"][j,:,:]
sla_decrease_step = paramVariete["slaPente"] * (data["sla"][j,:,:] - paramVariete["slaMin"])
# Modif du 10/07/2018, DeltaBiomasse neg si reallocation ne pas fair l'evol du SLA dans ces conditions
data["sla"][j:,:,:] = np.where(
(data["biomasseFeuille"][j,:,:] > 0),
np.where(
(data["deltaBiomasseFeuilles"][j,:,:] > 0),
#// (data["sla"][j,:,:] - paramVariete["slaPente"] * (data["sla"][j,:,:] - paramVariete["slaMin"])) * (data["biomasseFeuille"][j,:,:] - data["deltaBiomasseFeuilles"][j,:,:]) / data["biomasseFeuille"][j,:,:] + (paramVariete["slaMax"] + data["sla"][j,:,:])/2 * (data["deltaBiomasseFeuilles"][j,:,:] / data["biomasseFeuille"][j,:,:]),
(data["sla"][j,:,:] - sla_decrease_step) * ratio_old_leaf_biomass + (paramVariete["slaMax"] + data["sla"][j,:,:])/2 * ratio_new_leaf_biomass,
#//(data["sla"][j,:,:] - paramVariete["slaPente"] * (data["sla"][j,:,:] - paramVariete["slaMin"])) * (data["biomasseFeuille"][j,:,:] / data["biomasseFeuille"][j,:,:]),
(data["sla"][j,:,:] - sla_decrease_step) * ratio_old_leaf_biomass,
),
data["sla"][j,:,:],
)
data["sla"][j:,:,:] = np.where(
(data["biomasseFeuille"][j,:,:] > 0),
#// np.minimum(paramVariete["slaMin"], np.maximum(paramVariete["slaMax"], data["sla"][j,:,:])), # according to original
# according to ocelet version
np.minimum(
paramVariete["slaMax"],
np.maximum(
paramVariete["slaMin"],
data["sla"][j,:,:],
),
),
data["sla"][j,:,:],
)
return data
def calculate_leaf_area_index(j, data):
"""
Calculate the leaf area index (LAI) for a given time step.
If the number of growth phase (numPhase) is less than or equal to 1, the LAI is set to 0.
If the number of growth phase is between 2 and 6, the LAI is calculated as the product of
the leaf biomass (biomasseFeuille) and specific leaf area (sla).
If the number of growth phase is greater than 6, the LAI is set back to 0.
This function is adapted from the EvolLAIPhases procedure from the
milbilancarbone.pas and exmodules 1 & 2.pas file of the original Pascal
code.
Args:
timestep (int): The time step to calculate the LAI for.
data (xarray.Dataset): The xarray dataset that contains the relevant data.
Returns:
xarray.Dataset: The updated xarray dataset with the calculated LAI.
"""
data["lai"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] <= 1),
0,
np.where(
data["numPhase"][j,:,:] <= 6,
data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:],
0,
)
)
return data
def update_yield_during_filling_phase(j, data):
"""
This function updates the yield value during the filling phase.
During the filling phase (numPhase == 5), the yield is updated by
incrementing it with the sum of `deltaBiomasseAerienne` and `reallocation`,
bounded by 0 and `dRdtPot` (daily potential yield). The construction of yield
is done during phase 5 only, from the variation of aerial biomass and
reallocation, with a maximum of `dRdtPot`.
This function is adapted from the EvolDayRdtSarraV3 procedure from the
***bilancarbonesarra***, exmodules 1 & 2.pas file of the original Pascal
code.
Notes :
On tend vers le potentiel en fn du rapport des degresJours/sumDegresJours
pour la phase de remplissage. Frein sup fn du flux de sève estimé par le
rapport Tr/TrPot.
dRdtPot = RdtPotDuJour
Args:
j (int): The time step at which the calculation starts.
data (xarray.Dataset): The data that contains all variables.
Returns:
xarray.Dataset: The input data with updated yield values.
"""
data["rdt"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5),
data["rdt"][j,:,:] + np.minimum(data["dRdtPot"][j,:,:], np.maximum(0.0, data["deltaBiomasseAerienne"][j,:,:]) + data['reallocation'][j,:,:]),
data["rdt"][j,:,:],
)
return data
def BiomDensiteSarraV42(j, data, paramITK, paramVariete):
# depuis bilancarbonsarra.pas
if ~np.isnan(paramVariete["densOpti"]):
data["rdt"][j:,:,:] = (data["rdt"][j,:,:] / data["rapDensite"])
data["rdtPot"][j:,:,:] = (data["rdtPot"][j,:,:]/ data["rapDensite"])
data["biomasseRacinaire"][j:,:,:] = (data["biomasseRacinaire"][j,:,:] / data["rapDensite"])
data["biomasseTige"][j:,:,:] = (data["biomasseTige"][j,:,:] / data["rapDensite"])
data["biomasseFeuille"][j:,:,:] = (data["biomasseFeuille"][j,:,:] / data["rapDensite"])
data["biomasseAerienne"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:])
#? conflit avec fonction evolLAIphase ?
#data["lai"][j:,:,:] = data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:]
data["lai"][j:,:,:] = data["lai"][j:,:,:] / data["rapDensite"]
data["biomasseTotale"][j:,:,:] = (data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:])#[...,np.newaxis]
#data["biomasseTotale"][j:,:,:] = data["biomasseTotale"][j:,:,:] / data["rapDensite"]
return data
def BiomMcUBTSV3(j, data, paramITK):
"""
depuis bilancarbonsarra.pas
group 174
Pendant la croissance des cultures la d�gradation des r�sidusest calcul�e sans les UBT
Ici c'est pendant la saion s�che quand il n'y a des cultures pas de b�tes.
Sur le mulch dress� (Up) ou couch� Lit), on calcul sa d�gradation journali�re
sur les feuilles et les tiges en fn de coef KN (climat, termites...),
KI ingestion par les b�tes pression en UBT seulement pour les feuilles, KT (effet pi�tinement) qui va faire passer
du stade lev� en couch� et du stade couch� en ensevelissement pression en UBT
Par D�faut :
KNUp = 0.001 /jour
KNLit = 0.011
KN est soit une constante soit peut varier en fn climat (pas fait ref STEP)
KT = 0.003
KI = 0.005
NbUBT = 10 (zone Fakara)
"""
condition = (data["numPhase"][j,:,:] > 0)
# group 161
data["UBTCulture"][j:,:,:] = np.where(condition, 0, data["NbUBT"][j,:,:])#[...,np.newaxis]
# group 162
data["LitFeuille"][j:,:,:] = np.where(condition, data["LitFeuille"][j,:,:] + data["FeuilleUp"][j,:,:], data["LitFeuille"][j,:,:])#[...,np.newaxis]
# group 163
data["LitTige"][j:,:,:] = np.where(condition, data["LitTige"][j,:,:] + data["TigeUp"][j,:,:], data["LitTige"][j,:,:])#[...,np.newaxis]
# group 164
data["FeuilleUp"][j:,:,:] = np.where(condition, 0, data["FeuilleUp"][j,:,:])#[...,np.newaxis]
# group 165
data["TigeUp"][j:,:,:] = np.where(condition, 0, data["TigeUp"][j,:,:])#[...,np.newaxis]
# group 166
data["biomMc"][j:,:,:] = np.where(condition, data["LitFeuille"][j,:,:] + data["LitTige"][j,:,:], data["biomMc"][j,:,:])#[...,np.newaxis]
#// D�gradation des feuilles et tiges dress�es
# FeuilleUp := max(0, (FeuilleUp - FeuilleUp * KNUp - FeuilleUp * KI * UBTCulture - FeuilleUp * KT * UBTCulture));
# group 167
data["FeuilleUp"][j:,:,:] = np.maximum(
0,
data["FeuilleUp"][j,:,:] - data["FeuilleUp"][j,:,:] * paramITK["KNUp"] - data["FeuilleUp"][j,:,:] \
* paramITK["KI"] * data["UBTCulture"][j,:,:] - data["FeuilleUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
# group 168
# TigeUp := max(0, (TigeUp - TigeUp * KNUp - TigeUp * KT * UBTCulture));
data["TigeUp"][j:,:,:] = np.maximum(
0,
data["TigeUp"][j,:,:] - data["TigeUp"][j,:,:] * paramITK["KNUp"] - data["TigeUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
#// D�gradation des feuilles et tiges couch�es (liti�re)
# group 169
# LitFeuille := max(0, (LitFeuille - LitFeuille * KNLit - LitFeuille * KI * UBTCulture - LitFeuille * KT * UBTCulture));
data["LitFeuille"][j:,:,:] = np.maximum(
0,
data["LitFeuille"][j,:,:] - data["LitFeuille"][j,:,:] * paramITK["KNLit"] - data["LitFeuille"][j,:,:] * paramITK["KI"] \
* data["UBTCulture"][j,:,:] - data["LitFeuille"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
# group 170
# LitTige := max(0, (LitTige - LitTige * KNLit - LitTige * KT * UBTCulture));
data["LitTige"][j:,:,:] = np.maximum(
0,
data["LitTige"][j,:,:] - data["LitTige"][j,:,:] * paramITK["KNLit"] - data["LitTige"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
# group 171
#BiomMc := LitFeuille + LitTige;
data["biomMc"][j:,:,:] = (data["LitFeuille"][j,:,:] + data["LitTige"][j,:,:])#[...,np.newaxis]
# #// transfert dress� � liti�re effet pi�tinement
# LitFeuille := LitFeuille + FeuilleUp * KT * UBTCulture;
# group 172
data["LitFeuille"][j:,:,:] = (data["LitFeuille"][j,:,:] + data["FeuilleUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:])#[...,np.newaxis]
# LitTige := LitTige + TigeUp * KT * UBTCulture;
# group 173
data["LitTige"][j:,:,:] = (data["LitTige"][j,:,:] + data["TigeUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:])#[...,np.newaxis]
# // le 01/03 on consid�re que toutes les pailles et feuilles dressees sont couchees
# if (trunc(DayOfTheYear(DateEnCours)) = 61) then
# begin
# LitFeuille := LitFeuille + FeuilleUp;
# LitTige := LitTige + TigeUp;
# FeuilleUp := 0;
# TigeUp := 0;
# BiomMc := LitFeuille + LitTige;
# end;
return data
def MAJBiomMcSV3(data):
"""
groupe 182
A la Recolte, on calcul la part des biomasses qui restent sur place (Up), non r�colt�es
et la part qui est mise � terre (Liti�re) sur ce qui est laiss� sur place
On met a jour aussi la biomasse des liti�res pour les calculs effet mulch sue lr bilan hydrique
"""
# if (NumPhase =7) then
# begin
# groupe 175
# FeuilleUp := FeuilleUp + BiomasseFeuilles * (1-TxRecolte);
# groupe 176
# TigeUp := TigeUp + BiomasseTiges * (1-TxRecolte);
# groupe 177
# LitFeuille := LitFeuille + FeuilleUp * TxaTerre;
# groupe 178
# LitTige := LitTige + TigeUp * TxaTerre;
# groupe 179
# FeuilleUp := FeuilleUp * (1-TxaTerre);
# groupe 180
# TigeUp := TigeUp * (1-TxaTerre);
# // LitTige := LitTige + BiomMc;
# groupe 181
# BiomMC := LitFeuille + LitTige;
# { BiomasseFeuilles := 0;
# BiomasseTiges := 0;
return data
def estimate_critical_nitrogen_concentration(j, data):
# estimate critical nitrogen concentration from plant dry matter using the Justes et al (1994) relationship
data["Ncrit"][j,:,:] = 5.35 * (data["biomasseTotale"][j,:,:]/1000) ** (-0.44)
return data
Functions
BiomDensOptSarraV4
def BiomDensOptSarraV4(
j,
data,
paramITK
)
si densité plus faible alors on considére qu'il faut augmenter les biomasses, LAI etc
en regard de cette situation au niveau de chaque plante (car tout est rapporté é des kg/ha). Si elle est plus forte on ne change rien pour lors. Valeur fixe en ref au maés é déf en paramétre par variétésé rapDensite := Max(1, 70000/densite);
View Source
def BiomDensOptSarraV4(j, data, paramITK):
"""
si densité plus faible alors on considére qu'il faut augmenter les biomasses, LAI etc
en regard de cette situation au niveau de chaque plante (car tout est rapporté é des kg/ha).
Si elle est plus forte on ne change rien pour lors.
Valeur fixe en ref au maés é déf en paramétre par variétésé rapDensite := Max(1, 70000/densite);
"""
"""
if ~np.isnan(paramVariete["densOpti"]) :
paramITK["rapDensite"] = np.maximum(1,paramVariete["densOpti"]/paramITK["densite"])
data["rdt"][j,:,:] = data["rdt"][j,:,:] * paramITK["rapDensite"]
data["biomasseRacinaire"][j,:,:] = data["biomasseRacinaire"][j,:,:] * paramITK["rapDensite"]
data["biomasseTige"][j,:,:] = data["biomasseTige"][j,:,:] * paramITK["rapDensite"]
data["biomasseFeuille"][j,:,:] = data["biomasseFeuille"][j,:,:] * paramITK["rapDensite"]
data["biomasseAerienne"][j,:,:] = data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:]
data["lai"][j,:,:] = data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:]
data["biomasseTotale"][j,:,:] = data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:]
return data
"""
return data
BiomDensiteSarraV42
def BiomDensiteSarraV42(
j,
data,
paramITK,
paramVariete
)
View Source
def BiomDensiteSarraV42(j, data, paramITK, paramVariete):
# depuis bilancarbonsarra.pas
if ~np.isnan(paramVariete["densOpti"]):
data["rdt"][j:,:,:] = (data["rdt"][j,:,:] / data["rapDensite"])
data["rdtPot"][j:,:,:] = (data["rdtPot"][j,:,:]/ data["rapDensite"])
data["biomasseRacinaire"][j:,:,:] = (data["biomasseRacinaire"][j,:,:] / data["rapDensite"])
data["biomasseTige"][j:,:,:] = (data["biomasseTige"][j,:,:] / data["rapDensite"])
data["biomasseFeuille"][j:,:,:] = (data["biomasseFeuille"][j,:,:] / data["rapDensite"])
data["biomasseAerienne"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:])
#? conflit avec fonction evolLAIphase ?
#data["lai"][j:,:,:] = data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:]
data["lai"][j:,:,:] = data["lai"][j:,:,:] / data["rapDensite"]
data["biomasseTotale"][j:,:,:] = (data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:])#[...,np.newaxis]
#data["biomasseTotale"][j:,:,:] = data["biomasseTotale"][j:,:,:] / data["rapDensite"]
return data
BiomMcUBTSV3
def BiomMcUBTSV3(
j,
data,
paramITK
)
depuis bilancarbonsarra.pas
group 174
Pendant la croissance des cultures la d�gradation des r�sidusest calcul�e sans les UBT Ici c'est pendant la saion s�che quand il n'y a des cultures pas de b�tes. Sur le mulch dress� (Up) ou couch� Lit), on calcul sa d�gradation journali�re sur les feuilles et les tiges en fn de coef KN (climat, termites...), KI ingestion par les b�tes pression en UBT seulement pour les feuilles, KT (effet pi�tinement) qui va faire passer du stade lev� en couch� et du stade couch� en ensevelissement pression en UBT Par D�faut : KNUp = 0.001 /jour KNLit = 0.011 KN est soit une constante soit peut varier en fn climat (pas fait ref STEP) KT = 0.003 KI = 0.005 NbUBT = 10 (zone Fakara)
View Source
def BiomMcUBTSV3(j, data, paramITK):
"""
depuis bilancarbonsarra.pas
group 174
Pendant la croissance des cultures la d�gradation des r�sidusest calcul�e sans les UBT
Ici c'est pendant la saion s�che quand il n'y a des cultures pas de b�tes.
Sur le mulch dress� (Up) ou couch� Lit), on calcul sa d�gradation journali�re
sur les feuilles et les tiges en fn de coef KN (climat, termites...),
KI ingestion par les b�tes pression en UBT seulement pour les feuilles, KT (effet pi�tinement) qui va faire passer
du stade lev� en couch� et du stade couch� en ensevelissement pression en UBT
Par D�faut :
KNUp = 0.001 /jour
KNLit = 0.011
KN est soit une constante soit peut varier en fn climat (pas fait ref STEP)
KT = 0.003
KI = 0.005
NbUBT = 10 (zone Fakara)
"""
condition = (data["numPhase"][j,:,:] > 0)
# group 161
data["UBTCulture"][j:,:,:] = np.where(condition, 0, data["NbUBT"][j,:,:])#[...,np.newaxis]
# group 162
data["LitFeuille"][j:,:,:] = np.where(condition, data["LitFeuille"][j,:,:] + data["FeuilleUp"][j,:,:], data["LitFeuille"][j,:,:])#[...,np.newaxis]
# group 163
data["LitTige"][j:,:,:] = np.where(condition, data["LitTige"][j,:,:] + data["TigeUp"][j,:,:], data["LitTige"][j,:,:])#[...,np.newaxis]
# group 164
data["FeuilleUp"][j:,:,:] = np.where(condition, 0, data["FeuilleUp"][j,:,:])#[...,np.newaxis]
# group 165
data["TigeUp"][j:,:,:] = np.where(condition, 0, data["TigeUp"][j,:,:])#[...,np.newaxis]
# group 166
data["biomMc"][j:,:,:] = np.where(condition, data["LitFeuille"][j,:,:] + data["LitTige"][j,:,:], data["biomMc"][j,:,:])#[...,np.newaxis]
#// D�gradation des feuilles et tiges dress�es
# FeuilleUp := max(0, (FeuilleUp - FeuilleUp * KNUp - FeuilleUp * KI * UBTCulture - FeuilleUp * KT * UBTCulture));
# group 167
data["FeuilleUp"][j:,:,:] = np.maximum(
0,
data["FeuilleUp"][j,:,:] - data["FeuilleUp"][j,:,:] * paramITK["KNUp"] - data["FeuilleUp"][j,:,:] \
* paramITK["KI"] * data["UBTCulture"][j,:,:] - data["FeuilleUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
# group 168
# TigeUp := max(0, (TigeUp - TigeUp * KNUp - TigeUp * KT * UBTCulture));
data["TigeUp"][j:,:,:] = np.maximum(
0,
data["TigeUp"][j,:,:] - data["TigeUp"][j,:,:] * paramITK["KNUp"] - data["TigeUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
#// D�gradation des feuilles et tiges couch�es (liti�re)
# group 169
# LitFeuille := max(0, (LitFeuille - LitFeuille * KNLit - LitFeuille * KI * UBTCulture - LitFeuille * KT * UBTCulture));
data["LitFeuille"][j:,:,:] = np.maximum(
0,
data["LitFeuille"][j,:,:] - data["LitFeuille"][j,:,:] * paramITK["KNLit"] - data["LitFeuille"][j,:,:] * paramITK["KI"] \
* data["UBTCulture"][j,:,:] - data["LitFeuille"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
# group 170
# LitTige := max(0, (LitTige - LitTige * KNLit - LitTige * KT * UBTCulture));
data["LitTige"][j:,:,:] = np.maximum(
0,
data["LitTige"][j,:,:] - data["LitTige"][j,:,:] * paramITK["KNLit"] - data["LitTige"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:],
)#[...,np.newaxis]
# group 171
#BiomMc := LitFeuille + LitTige;
data["biomMc"][j:,:,:] = (data["LitFeuille"][j,:,:] + data["LitTige"][j,:,:])#[...,np.newaxis]
# #// transfert dress� � liti�re effet pi�tinement
# LitFeuille := LitFeuille + FeuilleUp * KT * UBTCulture;
# group 172
data["LitFeuille"][j:,:,:] = (data["LitFeuille"][j,:,:] + data["FeuilleUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:])#[...,np.newaxis]
# LitTige := LitTige + TigeUp * KT * UBTCulture;
# group 173
data["LitTige"][j:,:,:] = (data["LitTige"][j,:,:] + data["TigeUp"][j,:,:] * paramITK["KT"] * data["UBTCulture"][j,:,:])#[...,np.newaxis]
# // le 01/03 on consid�re que toutes les pailles et feuilles dressees sont couchees
# if (trunc(DayOfTheYear(DateEnCours)) = 61) then
# begin
# LitFeuille := LitFeuille + FeuilleUp;
# LitTige := LitTige + TigeUp;
# FeuilleUp := 0;
# TigeUp := 0;
# BiomMc := LitFeuille + LitTige;
# end;
return data
EvalAssimSarrahV4
def EvalAssimSarrahV4(
j,
data
)
data["parIntercepte"][j,:,:] = 0.5 * (1 - data["ltr"][j,:,:]) * data["rg"][j,:,:]
data["assimPot"][j:,:,:] = data["parIntercepte"][j,:,:] * data["conv"][j,:,:] * 10
data["assim"][j,:,:] = np.where( data["trPot"][j,:,:] > 0, data["assimPot"][j,:,:] * data["tr"][j,:,:] / data["trPot"][j,:,:], 0, )
View Source
def EvalAssimSarrahV4(j, data):
"""
data["parIntercepte"][j,:,:] = 0.5 * (1 - data["ltr"][j,:,:]) * data["rg"][j,:,:]
data["assimPot"][j:,:,:] = data["parIntercepte"][j,:,:] * data["conv"][j,:,:] * 10
data["assim"][j,:,:] = np.where(
data["trPot"][j,:,:] > 0,
data["assimPot"][j,:,:] * data["tr"][j,:,:] / data["trPot"][j,:,:],
0,
)
"""
return data
EvalFeuilleTigeSarrahV4
def EvalFeuilleTigeSarrahV4(
j,
data,
paramVariete
)
This function is a wrapper
It is adapted from the EvalFeuilleTigeSarrahV4 procedure from the bilancarbonsarra.pas file of the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
| paramVariete | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def EvalFeuilleTigeSarrahV4(j, data, paramVariete):
"""
This function is a wrapper
It is adapted from the EvalFeuilleTigeSarrahV4 procedure from the bilancarbonsarra.pas file
of the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
# data["deltaBiomasseFeuilles"][j:,:,:] = np.where(
# (data["numPhase"][j,:,:] > 1),
# data["biomasseFeuille"][j,:,:],
# data["deltaBiomasseFeuilles"][j,:,:],
# )
# if (data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] < 0)
data = update_leaf_biomass(j, data, paramVariete)
data = update_stem_biomass(j, data, paramVariete)
# if deltaBiomasseAerienne >= 0 and (numPhase <= 4 or numPhase <= phaseDevVeg)
data = update_bM_and_cM(j, data, paramVariete)
data = update_leaf_biomass_positive_delta_aboveground_biomass(j, data, paramVariete)
data = update_stem_biomass_positive_delta_aboveground_biomass(j, data, paramVariete)
# if deltaBiomasseAerienne > 0 and numPhase > 1
data = update_leaf_biomass_all_phases(j, data, paramVariete)
data = update_stem_biomass_all_phases(j, data, paramVariete)
# condition = (data["numPhase"][j,:,:] > 1)
# data["deltaBiomasseFeuilles"][j:,:,:] = np.where(
# (data["numPhase"][j,:,:] > 1),
# data["biomasseFeuille"][j,:,:] - data["deltaBiomasseFeuilles"][j,:,:],
# data["deltaBiomasseFeuilles"][j,:,:],
# )
# simpler formulation for updating the deltaBiomasseFeuilles
data["deltaBiomasseFeuilles"][j:,:,:] = data["biomasseFeuille"][j,:,:] - data["biomasseFeuille"][j-1,:,:]
data = update_aboveground_biomass_step_2(j, data)
return data
MAJBiomMcSV3
def MAJBiomMcSV3(
data
)
groupe 182
A la Recolte, on calcul la part des biomasses qui restent sur place (Up), non r�colt�es et la part qui est mise � terre (Liti�re) sur ce qui est laiss� sur place On met a jour aussi la biomasse des liti�res pour les calculs effet mulch sue lr bilan hydrique
View Source
def MAJBiomMcSV3(data):
"""
groupe 182
A la Recolte, on calcul la part des biomasses qui restent sur place (Up), non r�colt�es
et la part qui est mise � terre (Liti�re) sur ce qui est laiss� sur place
On met a jour aussi la biomasse des liti�res pour les calculs effet mulch sue lr bilan hydrique
"""
# if (NumPhase =7) then
# begin
# groupe 175
# FeuilleUp := FeuilleUp + BiomasseFeuilles * (1-TxRecolte);
# groupe 176
# TigeUp := TigeUp + BiomasseTiges * (1-TxRecolte);
# groupe 177
# LitFeuille := LitFeuille + FeuilleUp * TxaTerre;
# groupe 178
# LitTige := LitTige + TigeUp * TxaTerre;
# groupe 179
# FeuilleUp := FeuilleUp * (1-TxaTerre);
# groupe 180
# TigeUp := TigeUp * (1-TxaTerre);
# // LitTige := LitTige + BiomMc;
# groupe 181
# BiomMC := LitFeuille + LitTige;
# { BiomasseFeuilles := 0;
# BiomasseTiges := 0;
return data
adjust_for_sowing_density
def adjust_for_sowing_density(
j,
data,
paramVariete,
direction
)
This function translates the effect of sowing density on biomass and LAI.
This function is adapted from the BiomDensOptSarV42 and BiomDensiteSarraV42 procedures, from the bilancarbonsarra.pas original Pascal code.
Notes from CB : if density is lower than the optimal density, then we consider that we need to increase the biomass, LAI etc in regard of this situation at each plant level (because everything is related to kg/ha). If it is higher, it increases asymptotically.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
| paramITK | type | description | None |
| paramVariete | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def adjust_for_sowing_density(j, data, paramVariete, direction):
"""
This function translates the effect of sowing density on biomass and LAI.
This function is adapted from the BiomDensOptSarV42 and BiomDensiteSarraV42
procedures, from the bilancarbonsarra.pas original Pascal code.
Notes from CB :
if density is lower than the optimal density, then we consider that we need
to increase the biomass, LAI etc in regard of this situation at each plant
level (because everything is related to kg/ha). If it is higher, it
increases asymptotically.
Args:
j (_type_): _description_
data (_type_): _description_
paramITK (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
if direction == "in" :
if ~np.isnan(paramVariete["densOpti"]) :
data["rdt"][j:,:,:] = data["rdt"][j,:,:] * data["rapDensite"][j,:,:]
data["rdtPot"][j:,:,:] = (data["rdtPot"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseRacinaire"][j:,:,:] = (data["biomasseRacinaire"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseTige"][j:,:,:] = (data["biomasseTige"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseFeuille"][j:,:,:] = (data["biomasseFeuille"][j,:,:] * data["rapDensite"][j,:,:])
data["biomasseAerienne"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:])
data["lai"][j:,:,:] = (data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:])
data["biomasseTotale"][j:,:,:] = (data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:])
return data
if direction == "out":
if ~np.isnan(paramVariete["densOpti"]):
data["rdt"][j:,:,:] = (data["rdt"][j,:,:] / data["rapDensite"][j,:,:])
data["rdtPot"][j:,:,:] = (data["rdtPot"][j,:,:]/ data["rapDensite"][j,:,:])
data["biomasseRacinaire"][j:,:,:] = (data["biomasseRacinaire"][j,:,:] / data["rapDensite"][j,:,:])
data["biomasseTige"][j:,:,:] = (data["biomasseTige"][j,:,:] / data["rapDensite"][j,:,:])
data["biomasseFeuille"][j:,:,:] = (data["biomasseFeuille"][j,:,:] / data["rapDensite"][j,:,:])
data["biomasseAerienne"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:])
#? conflit avec fonction evolLAIphase ?
#data["lai"][j:,:,:] = data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:]
data["lai"][j:,:,:] = data["lai"][j:,:,:] / data["rapDensite"][j,:,:]
data["biomasseTotale"][j:,:,:] = (data["biomasseAerienne"][j,:,:] + data["biomasseRacinaire"][j,:,:])#[...,np.newaxis]
#data["biomasseTotale"][j:,:,:] = data["biomasseTotale"][j:,:,:] / data["rapDensite"]
return data
calculate_canopy_specific_leaf_area
def calculate_canopy_specific_leaf_area(
j,
data,
paramVariete
)
Calculate the specific leaf area (SLA) of the canopy.
If the leaf biomass is positive, and if we are at the transition day between
phases 1 and 2 (numPhase = 2 and changePhase = 1), then the SLA is set to
slaMax.
If the leaf biomass is positive and increasing (deltaBiomasseFeuilles is
positive), the SLA for existing leaves is calculated by reducing it by an
amount proportional to the current SLA, while the SLA for new leaves is
calculated as the average between SLA and slaMax. The SLA for the entire
canopy is then calculated as the weighted average of the SLAs for existing
and new leaves.
If there is no increase in leaf biomass (deltaBiomasseFeuilles is negative), only the SLA for existing leaves is calculated.
If the leaf biomass is negative, the SLA is unchanged.
The calculated SLA value is bounded between slaMin and slaMax.
This function is adapted from the EvalSlaSarrahV3 procedure in the
bilancarbonsarra.pas and exmodules 1 & 2.pas files of the original Pascal
code. This calculation method assumes that young leaves have a higher SLA
than old leaves and that the fraction of young leaves makes the canopy SLA
increase. The penteSLA parameter causes a general decrease in SLA
(penteSLA = relative decrease per day = fraction of difference between
slaMax and slaMin).
Expected parameters: SLAmax [0.001, 0.01] SLAmin [0.001, 0.01] penteSLA [0, 0.2] SLAini = SLAmax
This function estimates the specific leaf area (SLA) of the canopy.
First, if the leaf biomass is positive, if numPhase = 2 and changePhase = 1, which means we are at the transition day between phases 1 and 2, sla is set to be equal to slaMax.
Then, if the leaf biomass is positive, and if deltaBiomasseFeuilles is positive (meaning that the leaf biomass is increasing), SLA for already existing leaves is calculated by removing a value that is an affine function of SLA itself, and SLA for new leaves is calculated as the mean between SLA and slaMax ; then the SLA is calculated as the weighted mean of the two SLA values.
Logically, if there is no newly produced leaf biomass (deltaBiomasseFeuilles is negative), only the SLA for already existing leaves is calculated.
If biomasseFeuille is negative, SLA is unchanged.
Finally, if biomasseFeuille is positive, SLA value is bounded between slaMin and slaMax.
This function is adapted from the EvalSlaSarrahV3 procedure from the bilancarbonsarra.pas and exmodules 1 & 2.pas file of the original Pascal code. We note that multiple versions of the calculation methods have been used in the original procecure. We may want to go back to that if this function is problematic.
Notes : In this approach, it is assumed that young leaves have a higher SLA than old leaves. The fraction of young leaves makes the canopy SLA increase. The penteSLA parameter causes a general decrease in SLA (penteSLA = relative decrease per day = fraction of difference between SLAmax and SLAmin). This approach is known for legumes, but can also be adapted to other species.
Generic/expected parameters : SLAmax [0.001, 0.01] SLAmin [0.001, 0.01] penteSLA [0, 0.2] SLAini = SLAmax
Args: - j (int): The time step. - data (xarray.Dataset): The data for all variables. - paramVariete (dict): Parameters for the calculation.
Returns: - data (xarray.Dataset): The updated data with the calculated SLA.
View Source
def calculate_canopy_specific_leaf_area(j, data, paramVariete):
"""
Calculate the specific leaf area (SLA) of the canopy.
If the leaf biomass is positive, and if we are at the transition day between
phases 1 and 2 (numPhase = 2 and changePhase = 1), then the SLA is set to
`slaMax`.
If the leaf biomass is positive and increasing (deltaBiomasseFeuilles is
positive), the SLA for existing leaves is calculated by reducing it by an
amount proportional to the current SLA, while the SLA for new leaves is
calculated as the average between SLA and `slaMax`. The SLA for the entire
canopy is then calculated as the weighted average of the SLAs for existing
and new leaves.
If there is no increase in leaf biomass (deltaBiomasseFeuilles is negative),
only the SLA for existing leaves is calculated.
If the leaf biomass is negative, the SLA is unchanged.
The calculated SLA value is bounded between `slaMin` and `slaMax`.
This function is adapted from the EvalSlaSarrahV3 procedure in the
bilancarbonsarra.pas and exmodules 1 & 2.pas files of the original Pascal
code. This calculation method assumes that young leaves have a higher SLA
than old leaves and that the fraction of young leaves makes the canopy SLA
increase. The `penteSLA` parameter causes a general decrease in SLA
(penteSLA = relative decrease per day = fraction of difference between
`slaMax` and `slaMin`).
Expected parameters:
SLAmax [0.001, 0.01]
SLAmin [0.001, 0.01]
penteSLA [0, 0.2]
SLAini = SLAmax
This function estimates the specific leaf area (SLA) of the canopy.
First, if the leaf biomass is positive, if numPhase = 2 and changePhase = 1,
which means we are at the transition day between phases 1 and 2, sla is set
to be equal to slaMax.
Then, if the leaf biomass is positive, and if deltaBiomasseFeuilles is
positive (meaning that the leaf biomass is increasing), SLA for already
existing leaves is calculated by removing a value that is an affine function
of SLA itself, and SLA for new leaves is calculated as the mean between SLA
and slaMax ; then the SLA is calculated as the weighted mean of the two SLA
values.
Logically, if there is no newly produced leaf biomass (deltaBiomasseFeuilles
is negative), only the SLA for already existing leaves is calculated.
If biomasseFeuille is negative, SLA is unchanged.
Finally, if biomasseFeuille is positive, SLA value is bounded between slaMin
and slaMax.
This function is adapted from the EvalSlaSarrahV3 procedure from the
bilancarbonsarra.pas and exmodules 1 & 2.pas file of the original Pascal
code. We note that multiple versions of the calculation methods have been
used in the original procecure. We may want to go back to that if this
function is problematic.
Notes :
In this approach, it is assumed that young leaves have a higher SLA than old
leaves. The fraction of young leaves makes the canopy SLA increase. The
penteSLA parameter causes a general decrease in SLA (penteSLA = relative
decrease per day = fraction of difference between SLAmax and SLAmin). This
approach is known for legumes, but can also be adapted to other species.
Generic/expected parameters :
SLAmax [0.001, 0.01]
SLAmin [0.001, 0.01]
penteSLA [0, 0.2]
SLAini = SLAmax
Args:
- j (int): The time step.
- data (xarray.Dataset): The data for all variables.
- paramVariete (dict): Parameters for the calculation.
Returns:
- data (xarray.Dataset): The updated data with the calculated SLA.
"""
condition = (data["biomasseFeuille"][j,:,:] > 0) & \
(data["numPhase"][j,:,:] == 2) & \
(data["changePhase"][j,:,:] == 1)
data["sla"][j:,:,:] = np.where(
condition,
paramVariete["slaMax"],
data["sla"][j,:,:],
)
ratio_old_leaf_biomass = data["biomasseFeuille"][j-1,:,:] / data["biomasseFeuille"][j,:,:]
ratio_new_leaf_biomass = data["deltaBiomasseFeuilles"][j,:,:] / data["biomasseFeuille"][j,:,:]
sla_decrease_step = paramVariete["slaPente"] * (data["sla"][j,:,:] - paramVariete["slaMin"])
# Modif du 10/07/2018, DeltaBiomasse neg si reallocation ne pas fair l'evol du SLA dans ces conditions
data["sla"][j:,:,:] = np.where(
(data["biomasseFeuille"][j,:,:] > 0),
np.where(
(data["deltaBiomasseFeuilles"][j,:,:] > 0),
#// (data["sla"][j,:,:] - paramVariete["slaPente"] * (data["sla"][j,:,:] - paramVariete["slaMin"])) * (data["biomasseFeuille"][j,:,:] - data["deltaBiomasseFeuilles"][j,:,:]) / data["biomasseFeuille"][j,:,:] + (paramVariete["slaMax"] + data["sla"][j,:,:])/2 * (data["deltaBiomasseFeuilles"][j,:,:] / data["biomasseFeuille"][j,:,:]),
(data["sla"][j,:,:] - sla_decrease_step) * ratio_old_leaf_biomass + (paramVariete["slaMax"] + data["sla"][j,:,:])/2 * ratio_new_leaf_biomass,
#//(data["sla"][j,:,:] - paramVariete["slaPente"] * (data["sla"][j,:,:] - paramVariete["slaMin"])) * (data["biomasseFeuille"][j,:,:] / data["biomasseFeuille"][j,:,:]),
(data["sla"][j,:,:] - sla_decrease_step) * ratio_old_leaf_biomass,
),
data["sla"][j,:,:],
)
data["sla"][j:,:,:] = np.where(
(data["biomasseFeuille"][j,:,:] > 0),
#// np.minimum(paramVariete["slaMin"], np.maximum(paramVariete["slaMax"], data["sla"][j,:,:])), # according to original
# according to ocelet version
np.minimum(
paramVariete["slaMax"],
np.maximum(
paramVariete["slaMin"],
data["sla"][j,:,:],
),
),
data["sla"][j,:,:],
)
return data
calculate_leaf_area_index
def calculate_leaf_area_index(
j,
data
)
Calculate the leaf area index (LAI) for a given time step.
If the number of growth phase (numPhase) is less than or equal to 1, the LAI is set to 0. If the number of growth phase is between 2 and 6, the LAI is calculated as the product of the leaf biomass (biomasseFeuille) and specific leaf area (sla). If the number of growth phase is greater than 6, the LAI is set back to 0.
This function is adapted from the EvolLAIPhases procedure from the milbilancarbone.pas and exmodules 1 & 2.pas file of the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| timestep | int | The time step to calculate the LAI for. | None |
| data | xarray.Dataset | The xarray dataset that contains the relevant data. | None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The updated xarray dataset with the calculated LAI. |
View Source
def calculate_leaf_area_index(j, data):
"""
Calculate the leaf area index (LAI) for a given time step.
If the number of growth phase (numPhase) is less than or equal to 1, the LAI is set to 0.
If the number of growth phase is between 2 and 6, the LAI is calculated as the product of
the leaf biomass (biomasseFeuille) and specific leaf area (sla).
If the number of growth phase is greater than 6, the LAI is set back to 0.
This function is adapted from the EvolLAIPhases procedure from the
milbilancarbone.pas and exmodules 1 & 2.pas file of the original Pascal
code.
Args:
timestep (int): The time step to calculate the LAI for.
data (xarray.Dataset): The xarray dataset that contains the relevant data.
Returns:
xarray.Dataset: The updated xarray dataset with the calculated LAI.
"""
data["lai"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] <= 1),
0,
np.where(
data["numPhase"][j,:,:] <= 6,
data["biomasseFeuille"][j,:,:] * data["sla"][j,:,:],
0,
)
)
return data
calculate_maintainance_respiration
def calculate_maintainance_respiration(
j,
data,
paramVariete
)
This function calculates the maintenance respiration respMaint (in kg/ha/j in equivalent dry matter) of the plant.
The maintenance respiration is calculated by summing the maintenance respiration associated with total biomass and
leaves biomass. If the plant's growth phase is above 4 and there is no leaf biomass, respMaint is set to 0.
The calculation is based on the equation: coefficient_temp = 2^((average_temp - tempMaint) / 10) respiration = kRespMaint * biomass * coefficient_temp
where average_temp is the average temperature for the day, tempMaint is the maintenance temperature from
variety_params, kRespMaint is the maintenance respiration coefficient from variety_params, and biomass
is the total or leaf biomass.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | The time step of the calculation. | None |
| data | xarray.Dataset | The input data containing the variables tpMoy, biomasseTotale, biomasseFeuille, and numPhase. The output respMaint will also be stored in this dataset. |
None |
| variety_params | dict | The parameters related to the plant variety, containing the keys tempMaint and kRespMaint. |
None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The input data with the updated respMaint variable. |
View Source
def calculate_maintainance_respiration(j, data, paramVariete):
"""
This function calculates the maintenance respiration `respMaint` (in kg/ha/j in equivalent dry matter) of the plant.
The maintenance respiration is calculated by summing the maintenance respiration associated with total biomass and
leaves biomass. If the plant's growth phase is above 4 and there is no leaf biomass, `respMaint` is set to 0.
The calculation is based on the equation:
coefficient_temp = 2^((average_temp - tempMaint) / 10)
respiration = kRespMaint * biomass * coefficient_temp
where `average_temp` is the average temperature for the day, `tempMaint` is the maintenance temperature from
`variety_params`, `kRespMaint` is the maintenance respiration coefficient from `variety_params`, and `biomass`
is the total or leaf biomass.
Args:
j (int): The time step of the calculation.
data (xarray.Dataset): The input data containing the variables `tpMoy`, `biomasseTotale`, `biomasseFeuille`, and
`numPhase`. The output `respMaint` will also be stored in this dataset.
variety_params (dict): The parameters related to the plant variety, containing the keys `tempMaint` and
`kRespMaint`.
Returns:
xarray.Dataset: The input `data` with the updated `respMaint` variable.
"""
coefficient_temp = 2**((data["tpMoy"][j,:,:] - paramVariete["tempMaint"]) / 10)
resp_totale = paramVariete["kRespMaint"] * data["biomasseTotale"][j,:,:] * coefficient_temp
resp_feuille = paramVariete["kRespMaint"] * data["biomasseFeuille"][j,:,:] * coefficient_temp
data["respMaint"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 4) & (data["biomasseFeuille"][j,:,:]==0),
0,
resp_totale + resp_feuille,
)
return data
compute_rapDensite
def compute_rapDensite(
paramITK,
paramVariete
)
It basically calculates a correction factor (rapDensite).
This correction factor Is calculated with an equation of form
a + p * exp( -(x/(o / -log((1-a)/p) )) )
with a the densiteA parameter p the densiteP parameter$ x the actual crop density o the densOpti parameter
See https://www.wolframalpha.com/input?i=a+%2B+p+*+exp%28-%28x+%2F+%28+o%2F-+log%28%281+-+a%29%2F+p%29%29%29%29 for equation visualization.
This equation is probably too complex for the problem at hand.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
| paramITK | type | description | None |
| paramVariete | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def compute_rapDensite(paramITK, paramVariete):
"""
It basically calculates a correction factor (rapDensite).
This correction factor Is calculated with an equation of form
a + p * exp( -(x/(o / -log((1-a)/p) )) )
with a the densiteA parameter
p the densiteP parameter$
x the actual crop density
o the densOpti parameter
See
https://www.wolframalpha.com/input?i=a+%2B+p+*+exp%28-%28x+%2F+%28+o%2F-+log%28%281+-+a%29%2F+p%29%29%29%29
for equation visualization.
This equation is probably too complex for the problem at hand.
Args:
j (_type_): _description_
data (_type_): _description_
paramITK (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
rapDensite = paramVariete["densiteA"] + paramVariete["densiteP"] * np.exp(-(paramITK["densite"] / ( paramVariete["densOpti"]/- np.log((1 - paramVariete['densiteA'])/ paramVariete["densiteP"]))))
return rapDensite
condition_positive_delta_aboveground_biomass_all_phases
def condition_positive_delta_aboveground_biomass_all_phases(
j,
data
)
View Source
def condition_positive_delta_aboveground_biomass_all_phases(j, data):
#// condition = (data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] >= 0)
condition = (data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] > 0)
return condition
condition_positive_delta_biomass
def condition_positive_delta_biomass(
j,
data,
paramVariete
)
View Source
def condition_positive_delta_biomass(j, data, paramVariete):
condition = (data["numPhase"][j,:,:] > 1) & \
(data["deltaBiomasseAerienne"][j,:,:] >= 0) & \
((data["numPhase"][j,:,:] <= 4) | (data["numPhase"][j,:,:] <= paramVariete["phaseDevVeg"]))
# (data["numPhase"][j,:,:] <= 4)
return condition
estimate_KAssim
def estimate_KAssim(
j,
data,
paramVariete
)
This function calculates the conversion factor KAssim, which is used to convert assimilates into biomass.
The value of KAssim depends on the phase of the crop.
The conversion factor is calculated based on a lookup table that maps crop phases to values. The crop phase is
determined by the numPhase field in the data argument, and the corresponding KAssim value is set in the
KAssim field of the data argument.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | An integer index specifying the time step. | None |
| data | xarray.Dataset | A dataset containing the variables used in the calculation of KAssim. The dataset should include the fields numPhase, sdj, seuilTemp PhasePrec, and seuilTemp PhaseSuivante. TheKAssim field of the dataset will be updated by this function. |
None |
| paramVariete | dict | A dictionary of parameters. It should include the fields txAssimBVP, txAssimMatu1,and txAssimMatu2. |
None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The updated data dataset, with the KAssim field set to the calculated values. |
View Source
def estimate_KAssim(j, data, paramVariete):
"""
This function calculates the conversion factor `KAssim`, which is used to convert assimilates into biomass.
The value of `KAssim` depends on the phase of the crop.
The conversion factor is calculated based on a lookup table that maps crop phases to values. The crop phase is
determined by the `numPhase` field in the `data` argument, and the corresponding `KAssim` value is set in the
`KAssim` field of the `data` argument.
Args:
j (int): An integer index specifying the time step.
data (xarray.Dataset): A dataset containing the variables used in the calculation of `KAssim`. The dataset
should include the fields `numPhase`, `sdj`, `seuilTemp PhasePrec`, and `seuilTemp PhaseSuivante`. The
`KAssim` field of the dataset will be updated by this function.
paramVariete (dict): A dictionary of parameters. It should include the fields `txAssimBVP`, `txAssimMatu1`,
and `txAssimMatu2`.
Returns:
xarray.Dataset: The updated `data` dataset, with the `KAssim` field set to the calculated values.
"""
phase_equivalences = {
2: 1,
3: paramVariete['txAssimBVP'],
4: paramVariete['txAssimBVP'],
#! replacing sommeDegresJourPhasePrec with seuilTempPhasePrec
#// 5: paramVariete["txAssimBVP"] + (data['sdj'][j,:,:] - data['sommeDegresJourPhasePrec'][j,:,:]) * (paramVariete['txAssimMatu1'] - paramVariete['txAssimBVP']) / (data['seuilTempPhaseSuivante'][j,:,:] - data['sommeDegresJourPhasePrec'][j,:,:]),
5: paramVariete["txAssimBVP"] + (data['sdj'][j,:,:] - data['seuilTempPhasePrec'][j,:,:]) * (paramVariete['txAssimMatu1'] - paramVariete['txAssimBVP']) / (data['seuilTempPhaseSuivante'][j,:,:] - data['seuilTempPhasePrec'][j,:,:]),
#// 6: paramVariete["txAssimMatu1"] + (data["sdj"][j,:,:] - data["sommeDegresJourPhasePrec"][j,:,:]) * (paramVariete["txAssimMatu2"] - paramVariete["txAssimMatu1"]) / (data["seuilTempPhaseSuivante"][j,:,:] - data["sommeDegresJourPhasePrec"][j,:,:]),
6: paramVariete["txAssimMatu1"] + (data["sdj"][j,:,:] - data["seuilTempPhasePrec"][j,:,:]) * (paramVariete["txAssimMatu2"] - paramVariete["txAssimMatu1"]) / (data["seuilTempPhaseSuivante"][j,:,:] - data["seuilTempPhasePrec"][j,:,:]),
}
for phase in range(2,7):
data["KAssim"][j:,:,:] = np.where(
data["numPhase"][j,:,:] == phase,
phase_equivalences[phase],
data["KAssim"][j,:,:],
)
return data
estimate_conv
def estimate_conv(
j,
data,
paramVariete
)
This function calculates the conversion of assimilates into biomass.
The conversion factor is determined by multiplying the KAssim value, which
is dependent on the phase of the crop, with the conversion rate (txConversion)
specified in the paramVariete argument.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | The starting index of the calculation | None |
| data | dict | A dictionary containing information on the crop growth, including the phase of the crop and the KAssim value. |
None |
| paramVariete | dict | A dictionary containing parameters relevant to the crop growth, including the conversion rate. |
None |
Returns:
| Type | Description |
|---|---|
| dict | The input data dictionary with the calculated "conv" value added. |
View Source
def estimate_conv(j,data,paramVariete):
"""
This function calculates the conversion of assimilates into biomass.
The conversion factor is determined by multiplying the KAssim value, which
is dependent on the phase of the crop, with the conversion rate (txConversion)
specified in the `paramVariete` argument.
Args:
j (int): The starting index of the calculation
data (dict): A dictionary containing information on the crop growth, including
the phase of the crop and the KAssim value.
paramVariete (dict): A dictionary containing parameters relevant to the crop
growth, including the conversion rate.
Returns:
dict: The input `data` dictionary with the calculated "conv" value added.
"""
data["conv"][j:,:,:] = (data["KAssim"][j,:,:] * paramVariete["txConversion"])
return data
estimate_critical_nitrogen_concentration
def estimate_critical_nitrogen_concentration(
j,
data
)
View Source
def estimate_critical_nitrogen_concentration(j, data):
# estimate critical nitrogen concentration from plant dry matter using the Justes et al (1994) relationship
data["Ncrit"][j,:,:] = 5.35 * (data["biomasseTotale"][j,:,:]/1000) ** (-0.44)
return data
estimate_kcp
def estimate_kcp(
j,
data,
paramVariete
)
Estimate the kcp coefficient based on the maximum crop coefficient kcMax and plant cover ltr.
The computation of kcp is based on the EvolKcpKcIni procedure from the biomasse.pas and exmodules 1 & 2.pas files of the original PASCAL code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | The starting index for updating kcp in the data dataset. |
None |
| data | xarray.Dataset | A dataset containing the data used in the computation of kcp. The dataset should contain the following variables:- 'numPhase': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the number of phases in the crop cycle. - 'kcp': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the coefficient of crop growth. - 'ltr': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the plant cover. |
None |
| paramVariete | dict | A dictionary containing the parameters for estimating kcp. The dictionary should contain the following key:- 'kcMax': A float, representing the maximum crop coefficient. |
None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The updated data dataset with the new kcp values. |
View Source
def estimate_kcp(j, data, paramVariete):
"""
Estimate the kcp coefficient based on the maximum crop coefficient `kcMax` and plant cover `ltr`.
The computation of `kcp` is based on the EvolKcpKcIni procedure from the biomasse.pas and exmodules 1 & 2.pas files of the original PASCAL code.
Args:
j (int): The starting index for updating `kcp` in the `data` dataset.
data (xarray.Dataset): A dataset containing the data used in the computation of `kcp`. The dataset should contain the following variables:
- 'numPhase': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the number of phases in the crop cycle.
- 'kcp': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the coefficient of crop growth.
- 'ltr': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the plant cover.
paramVariete (dict): A dictionary containing the parameters for estimating `kcp`. The dictionary should contain the following key:
- 'kcMax': A float, representing the maximum crop coefficient.
Returns:
xarray.Dataset: The updated `data` dataset with the new `kcp` values.
"""
data["kcp"][j:,:,:] = np.where(
data["numPhase"][j,:,:] >= 1,
np.maximum(0.3, paramVariete["kcMax"] * (1 - data["ltr"][j,:,:])),
data["kcp"][j,:,:],
)
return data
estimate_ltr
def estimate_ltr(
j,
data,
paramVariete
)
Estimate the fraction of radiation transmitted to the soil ltr based on the leaf area index lai.
ltr is used as a proxy for plant covering of the soil in the water balance calculation, where 1 represents no plant cover and 0 represents full plant cover. ltr is computed as an exponential decay function of lai with a decay coefficient kdf.
This function is adapted from the EvalLtr procedure from the biomasse.pas and exmodules 1 & 2.pas files of the original PASCAL code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | The starting index for updating ltr in the data dataset. |
None |
| data | xarray.Dataset | A dataset containing the data used in the computation of ltr. The dataset should contain the following variables:- 'lai': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the leaf area index. - 'ltr': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the fraction of radiation transmitted to the soil. |
None |
| paramVariete | dict | A dictionary containing the parameters for estimating ltr. The dictionary should contain the following key:- 'kdf': A float, representing the decay coefficient for ltr. |
None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The updated data dataset with the new ltr values. |
View Source
def estimate_ltr(j, data, paramVariete):
"""
Estimate the fraction of radiation transmitted to the soil `ltr` based on the leaf area index `lai`.
`ltr` is used as a proxy for plant covering of the soil in the water balance calculation, where 1 represents no plant cover and 0 represents full plant cover. `ltr` is computed as an exponential decay function of `lai` with a decay coefficient `kdf`.
This function is adapted from the EvalLtr procedure from the biomasse.pas and exmodules 1 & 2.pas files of the original PASCAL code.
Args:
j (int): The starting index for updating `ltr` in the `data` dataset.
data (xarray.Dataset): A dataset containing the data used in the computation of `ltr`. The dataset should contain the following variables:
- 'lai': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the leaf area index.
- 'ltr': A 3-dimensional data variable with shape (num_timesteps, num_rows, num_columns), representing the fraction of radiation transmitted to the soil.
paramVariete (dict): A dictionary containing the parameters for estimating `ltr`. The dictionary should contain the following key:
- 'kdf': A float, representing the decay coefficient for `ltr`.
Returns:
xarray.Dataset: The updated `data` dataset with the new `ltr` values.
"""
# group 80
data["ltr"][j:,:,:] = np.exp(-paramVariete["kdf"] * data["lai"][j,:,:])
return data
estimate_reallocation
def estimate_reallocation(
j,
data,
paramVariete
)
Estimate the daily biomass reallocation between stem and leaves.
This function computes the daily biomass reallocation between stem and leaves for the plant. The computation only occurs when the plant is in phase 5. The amount of biomass that can be reallocated is estimated as follows:
-
The difference between the potential yield delta and the aboveground biomass delta, bound by 0, is calculated and referred to as manqueAssim. manqueAssim represents the daily variation in biomass that remains after the plant has built its aboveground biomass.
-
The reallocation is computed as the minimum of the product of manqueAssim and the reallocation rate and the difference between the leaf biomass and 30, also bound by 0. The value of 30 is an arbitrary threshold which ensures that reallocation is 0 if the leaf biomass is below 30. If the leaf biomass is above 30, reallocation is bounded by biomasseFeuille - 30.
If the plant is not in phase 5, reallocation is set to 0.
This function is based on the EvalReallocationSarrahV3 procedure from the bilancarbonsarra.pas and exmodules 1 & 2.pas files from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | Current time step of the simulation. | None |
| data | xarray.Dataset | The dataset containing all the simulation data. | None |
| paramVariete | dict | A dictionary containing the parameters for the simulation. | None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The updated dataset with the reallocation values. |
View Source
def estimate_reallocation(j, data, paramVariete):
"""
Estimate the daily biomass reallocation between stem and leaves.
This function computes the daily biomass reallocation between stem and leaves for the plant. The computation
only occurs when the plant is in phase 5. The amount of biomass that can be reallocated is estimated as
follows:
1. The difference between the potential yield delta and the aboveground biomass delta, bound by 0, is
calculated and referred to as manqueAssim. manqueAssim represents the daily variation in biomass that
remains after the plant has built its aboveground biomass.
2. The reallocation is computed as the minimum of the product of manqueAssim and the reallocation rate and
the difference between the leaf biomass and 30, also bound by 0. The value of 30 is an arbitrary
threshold which ensures that reallocation is 0 if the leaf biomass is below 30. If the leaf biomass is
above 30, reallocation is bounded by biomasseFeuille - 30.
If the plant is not in phase 5, reallocation is set to 0.
This function is based on the EvalReallocationSarrahV3 procedure from the bilancarbonsarra.pas and
exmodules 1 & 2.pas files from the original Pascal code.
Args:
j (int): Current time step of the simulation.
data (xarray.Dataset): The dataset containing all the simulation data.
paramVariete (dict): A dictionary containing the parameters for the simulation.
Returns:
xarray.Dataset: The updated dataset with the reallocation values.
"""
condition = (data["numPhase"][j,:,:] == 5)
data["manqueAssim"][j:,:,:] = np.where(
condition,
np.maximum(0, (data["dRdtPot"][j,:,:] - np.maximum(0.0, data["deltaBiomasseAerienne"][j,:,:]))),
0,
)
data["reallocation"][j:,:,:] = np.where(
condition,
np.minimum(
data["manqueAssim"][j,:,:] * paramVariete["txRealloc"],
np.maximum(0.0, data["biomasseFeuille"][j,:,:] - 30),
),
0,
)
return data
initialize_simulation
def initialize_simulation(
data,
grid_width,
grid_height,
duration,
paramVariete,
paramITK,
date_start
)
This function initializes variables related to crop growth in the data
xarray dataset. As the rain is the first variable to be initialized in the data xarray dataset, its dimensions are used to initialize the other variables.

This code has been adapted from the original InitiationCulture procedure, from the MilBilanCarbone.pas code of the
SARRA model.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| data | type | description grid_width (type): description | None |
| grid_height | type | description duration (type): description | None |
| paramVariete | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def initialize_simulation(data, grid_width, grid_height, duration, paramVariete, paramITK, date_start):
"""
This function initializes variables related to crop growth in the data
xarray dataset. As the rain is the first variable to be initialized in the
data xarray dataset, its dimensions are used to initialize the other
variables.

This code has been adapted from the original InitiationCulture procedure, from the `MilBilanCarbone.pas` code of the
SARRA model.
Args:
data (_type_): _description_ grid_width (_type_): _description_
grid_height (_type_): _description_ duration (_type_): _description_
paramVariete (_type_): _description_
Returns:
_type_: _description_
"""
### variables to be initialized with values from parameters
# from paramVariete : maximum daily thermal time (°C.j) -> #? unused ?
#// data["sommeDegresJourMaximale"] = (data["rain"].dims, np.full(
#// (duration, grid_width, grid_height),
#// (paramVariete["SDJLevee"] + paramVariete["SDJBVP"] + paramVariete["SDJRPR"] + paramVariete["SDJMatu1"] + paramVariete["SDJMatu2"])
#// ))
#// data["sommeDegresJourMaximale"].attrs = {"units":"°C.j", "long_name":"Maximum thermal time"}
# from paramITK : sowing date
data["sowing_date"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), (paramITK["DateSemis"] - date_start).days))
# from paramITK : automatic irrigation indicator
data["irrigAuto"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), paramITK["irrigAuto"]))
data["irrigAuto"].attrs = {"units":"binary", "long_name":"automatic irrigation indicator"}
####### variables qui viennent de initplotMc
# Initial biomass of crop residues (mulch) (kg/ha)
# Biomasse initiale des résidus de culture (mulch) (kg/ha)
# BiomMc := BiomIniMc;
data["biomMc"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), paramITK["biomIniMc"]))
data["biomMc"].attrs = {"units": "kg/ha", "long_name": "Initial biomass of crop residues (mulch)"}
data["biomMc"] = data["biomMc"].astype("float32")
# ?
# StSurf := StockIniSurf;
# data["stSurf"] = np.full((grid_width, grid_height, duration), paramTypeSol["stockIniSurf"])
# ?
# Ltr := 1;
data["ltr"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), 1.0))
data["ltr"] = data["ltr"].astype("float32")
# Initial biomass of stem residues as litter (kg/ha)
# Biomasse initiale des résidus de tiges sous forme de litière (kg/ha)
# LitTiges := BiomIniMc;
data["LitTige"] = (data["rain"].dims, np.full((duration, grid_width, grid_height), paramITK["biomIniMc"]))
data["LitTige"].attrs = {"units": "kg/ha", "long_name": "Initial biomass of stem residues as litter"}
data["LitTige"] = data["LitTige"].astype("float32")
####### fin variables qui viennent de initplotMc
####### variables eau depuis InitPlotMc
# Initializes variables related to crop residues boimass (mulch) in the data
# xarray dataset. This code has been adapted from the original InitPlotMc
# procedure, Bileau.pas code. Comments with tab indentation are from the
# original code. As the rain is the first variable to be initialized in the
# data xarray dataset, its dimensions are used to initialize the other
# variables.
# Soil maximum water storage capacity (mm)
# Capacité maximale de la RU (mm)
# StRurMax := Ru * ProfRacIni / 1000;
#! renaming stRurMax with root_tank_capacity
#// data["stRurMax"] = data["ru"] * paramITK["profRacIni"] / 1000
data["root_tank_capacity"] = (data["rain"].dims, np.repeat(np.array(data["ru"] * paramITK["profRacIni"] / 1000)[np.newaxis,:,:], duration, axis=0))
#// data["stRurMax"].attrs = {"units": "mm", "long_name": "Soil maximum water storage capacity"}
data["root_tank_capacity"].attrs = {"units": "mm", "long_name": "Soil maximum water storage capacity"}
# Maximum water capacity of surface tank (mm)
# Reserve utile de l'horizon de surface (mm)
# RuSurf := EpaisseurSurf / 1000 * Ru;
#! renaming ruSurf with surface_tank_capacity
#// data["ruSurf"] = data["epaisseurSurf"] / 1000 * data["ru"]
data["surface_tank_capacity"] = data["epaisseurSurf"] / 1000 * data["ru"]
#// data["ruSurf"].attrs = {"units": "mm", "long_name": "Maximum water capacity of surface tank"}
data["surface_tank_capacity"].attrs = {"units": "mm", "long_name": "Maximum water capacity of surface tank"}
# ?
# // PfTranspi := EpaisseurSurf * HumPf;
# // StTot := StockIniSurf - PfTranspi/2 + StockIniProf;
# StTot := StockIniSurf + StockIniProf;
# data["stTot"] = np.full((grid_width, grid_height, duration), (paramTypeSol["stockIniSurf"] + paramTypeSol["stockIniProf"]))
#! modifié pour faire correspondre les résultats de simulation, à remettre en place pour un calcul correct dès que possible
# data["stTot"] = np.full((grid_width, grid_height, duration), (paramTypeSol["stockIniProf"]))
#! renaming stTot to total_tank_stock
#// data["stTot"] = data["stockIniProf"]
#//data["total_tank_stock"] = data["stockIniProf"]
#! coorecting total_tank_stock initialization as it did not have the time dimensions that are required as stock evolves through time
data["total_tank_stock"] = (data["rain"].dims, np.repeat(np.array(data["stockIniProf"])[np.newaxis,:,:], duration, axis=0))
#// data["stTot"].attrs = {"units": "mm", "long_name": "?"}
data["total_tank_stock"].attrs = {"units": "mm", "long_name": "?"}
# Soil maximal depth (mm)
# Profondeur maximale de sol (mm)
# ProfRU := EpaisseurSurf + EpaisseurProf;
#! data["profRu"] = data["epaisseurProf"] + data["epaisseurSurf"]
#! data["profRu"].attrs = {"units": "mm", "long_name": "Soil maximal depth"}
# déplacé dans l'initialisation du sol
# Maximum water capacity to humectation front (mm)
# Quantité d'eau maximum jusqu'au front d'humectation (mm)
# // modif 10/06/2015 resilience stock d'eau
# // Front d'humectation egal a RuSurf trop de stress initial
# // Hum := max(StTot, StRurMax);
# Hum := max(RuSurf, StRurMax);
# // Hum mis a profRuSurf
# Hum := max(StTot, Hum);
data["humectation_front"] = (data["rain"].dims, np.full((duration, grid_width, grid_height),
np.maximum(
np.maximum(
#! renaming ruSurf with surface_tank_capacity
#// data["ruSurf"],
data["surface_tank_capacity"].expand_dims({"time":duration}),
#! renaming stRurMax with root_tank_capacity
#// data["stRurMax"],
data["root_tank_capacity"],
),
#! renaming stTot with total_tank_stock
#// data["stTot"],
data["total_tank_stock"],
)
))
data["humectation_front"].attrs = {"units": "mm", "long_name": "Maximum water capacity to humectation front"}
# Previous value for Maximum water capacity to humectation front (mm)
# HumPrec := Hum;
data["previous_humectation_front"] = data["humectation_front"]
# ?
# StRurPrec := 0;
# Previous value for stTot
# StRurMaxPrec := 0;
# //modif 10/06/2015 resilience stock d'eau
#! renaming stTot with total_tank_stock
#! renaminog stRuPrec with previous_total_tank_stock
#// data["stRuPrec"] = data["stTot"]
data["previous_total_tank_stock"] = data["total_tank_stock"]
####### fin variables eau depuis InitPlotMc
# depuis meteo.pas
kpar = 0.5
data["par"] = kpar * data["rg"]
data["par"].attrs = {"units":"MJ/m2", "long_name":"par"}
# crop density
if ~np.isnan(paramVariete["densOpti"]) :
data["rapDensite"] = data["rain"] * 0 + compute_rapDensite(paramITK, paramVariete)
data["rapDensite"].attrs = {"units":"none", "long_name":"sowing density adjustement factor"}
# initialize variables with values at 0
variables = variable_dict()
for variable in variables :
data[variable] = (data["rain"].dims, np.zeros(shape=(duration, grid_width, grid_height)))
data[variable].attrs = {"units":variables[variable][1], "long_name":variables[variable][0]}
data[variable] = data[variable].astype("float32")
return data
update_aboveground_biomass
def update_aboveground_biomass(
j,
data,
paramVariete
)
Update the aboveground biomass of the plant.
The aboveground biomass is either updated based on a linear function of the total biomass, if the plant is in phase 2, 3, or 4, or incremented with the total biomass delta if the plant is in any other phase.
This function is based on the EvolBiomAeroSarrahV3 procedure, of the bilancarbonsarra, exmodules 1 & 2.pas file from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | The current iteration step in the simulation. | None |
| data | xarray.Dataset | The simulation data, including the current phase of the plant and various biomass values. | None |
| paramVariete | dict | The parameters of the plant variety. | None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The updated simulation data, including the updated aboveground biomass and delta aboveground biomass. |
View Source
def update_aboveground_biomass(j, data, paramVariete):
"""
Update the aboveground biomass of the plant.
The aboveground biomass is either updated based on a linear function of the total biomass, if the plant is in phase 2, 3, or 4, or incremented with the total biomass delta if the plant is in any other phase.
This function is based on the EvolBiomAeroSarrahV3 procedure, of the
***bilancarbonsarra***, exmodules 1 & 2.pas file from the original Pascal
code.
Args:
j (int): The current iteration step in the simulation.
data (xarray.Dataset): The simulation data, including the current phase of the plant and various biomass values.
paramVariete (dict): The parameters of the plant variety.
Returns:
xarray.Dataset: The updated simulation data, including the updated aboveground biomass and delta aboveground biomass.
"""
#// data["deltaBiomasseAerienne"][j:,:,:] = np.copy(data["biomasseAerienne"][j,:,:])
data["biomasseAerienne"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] >= 2) & (data["numPhase"][j,:,:] <= 4),
np.minimum(0.9, paramVariete["aeroTotPente"] * data["biomasseTotale"][j,:,:] + paramVariete["aeroTotBase"]) * data["biomasseTotale"][j,:,:],
data["biomasseAerienne"][j,:,:] + data["deltaBiomasseTotale"][j,:,:],
)
#//data["deltaBiomasseAerienne"][j:,:,:] = (data["biomasseAerienne"][j,:,:] - data["deltaBiomasseAerienne"][j,:,:])#[...,np.newaxis]
data["deltaBiomasseAerienne"][j:,:,:] = data["biomasseAerienne"][j,:,:] - data["biomasseAerienne"][j-1,:,:]
return data
update_aboveground_biomass_step_2
def update_aboveground_biomass_step_2(
j,
data
)
summary
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_aboveground_biomass_step_2(j, data):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseAerienne"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 1),
data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:] + data["rdt"][j,:,:],
data["biomasseAerienne"][j,:,:],
)
return data
update_assim
def update_assim(
j,
data
)
This function updates assim. If trPot (potential transpiration from the
plant, mm) is greater than 0, then assim equals assimPot, multiplied by the ratio of effective transpiration over potential transpiration.
If potential transpiration is null, then assim is null as well.
Is it adapted from the EvalAssimSarraV42 procedure, of the bilancarbonsarra.pas file from the original Pascal code
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_assim(j, data):
"""
This function updates assim. If trPot (potential transpiration from the
plant, mm) is greater than 0, then assim equals assimPot, multiplied by the
ratio of effective transpiration over potential transpiration.
If potential transpiration is null, then assim is null as well.
Is it adapted from the EvalAssimSarraV42 procedure, of the
bilancarbonsarra.pas file from the original Pascal code
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["assim"][j,:,:] = np.where(
data["trPot"][j,:,:] > 0,
data["assimPot"][j,:,:] * data["tr"][j,:,:] / data["trPot"][j,:,:],
0,
)
return data
update_assimPot
def update_assimPot(
j,
data,
paramVariete,
paramITK
)
Update the assimPot value based on the intensification level (NI).
If the intensification level NI is defined in paramITK, the conversion rate txConversion is computed using a formula based on NIYo, NIp, LGauss, and AGauss. If NI is not defined, assimPot is updated using conv, which is updated in the estimate_conv function using the variables KAssim and txConversion.
When NI parameter is used (from to 4), conversion rate txConversion is computed using the following formula : NIYo + NIp * (1-exp(-NIp * NI)) - (exp(-0.5((NI - LGauss)/AGauss) (NI- LGauss)/AGauss))/(AGauss*2.506628274631)
This function is adapted from the EvalAssimSarraV42 procedure in the bilancarbonsarra.pas file of the original Pascal code.
Note from CB : correction of the conversion rate depending on the intensification level
notes from CB reharding the EvalAssimSarraV42 procedure :
Modif du 04/03/2021 : Prise en compte en plus de la densit� de semis de l'effet niveau d'intensification NI NI = 1 quand on est � l'optimum du niveau d'intensification. Dans le cas de situation contr�l� c'est la fertilit� qui est la clef principale en prenant en r�f�rence la qt� d'azote (�quivalent phosphore...) optimum Il peut aller � 0 ou �tre sup�rieur � 1 si situation sur optimum, ie un peu plus de rdt mais � cout trop �lev�... On �value un nouveau tx de conversion en fn du Ni au travers d'une double �quation : asympote x gaussienne invers�e Et d'un NI d�fini en fn du sc�nario de simulation ou des donn�es observ�es. NIYo = D�calage en Y de l'asymptote NIp = pente de l'asymptote LGauss = Largeur de la Guaussienne AGauss = Amplitude de la Guaussienne
Conversion qui est la valeur du taux de conversion en situation optimum n'a plus besoin d'�tre utilis� sinon dans la calibration des param�tres de cette �quation en absence de donn�es sur ces param�tres on ne met aucune valeur � NI CF fichier ex IndIntensite_txConv_eq.xls}
Args:
- j (int): An index that represents the current iteration.
- data (dict): A dictionary containing data arrays with the following keys:
- "assimPot" (np.ndarray): An array representing the potential assimilation rate.
- "par" (np.ndarray): An array representing photosynthetically active radiation.
- "lai" (np.ndarray): An array representing the leaf area index.
- "conv" (np.ndarray): An array representing the conversion rate.
- paramVariete (dict): A dictionary containing parameters for the computation of the conversion rate, including:
- "txConversion" (float): The conversion rate.
- "NIYo" (float): The shift in the Y-axis of the asymptote.
- "NIp" (float): The slope of the asymptote.
- "LGauss" (float): The width of the Gaussian curve.
- "AGauss" (float): The amplitude of the Gaussian curve.
- "kdf" (float): The constant used in the computation of assimPot.
- paramITK (dict): A dictionary containing the intensification level NI (float).
Returns:
- data (dict): The input data dictionary with the updated "assimPot" array.
View Source
def update_assimPot(j, data, paramVariete, paramITK):
"""
Update the assimPot value based on the intensification level (NI).
If the intensification level `NI` is defined in `paramITK`, the conversion rate `txConversion` is computed using a formula based on `NIYo`, `NIp`, `LGauss`, and `AGauss`. If `NI` is not defined, `assimPot` is updated using `conv`, which is updated in the `estimate_conv` function using the variables `KAssim` and `txConversion`.
When NI parameter is used (from to 4), conversion rate txConversion
is computed using the following formula :
NIYo + NIp * (1-exp(-NIp * NI)) - (exp(-0.5*((NI - LGauss)/AGauss)* (NI- LGauss)/AGauss))/(AGauss*2.506628274631)
This function is adapted from the `EvalAssimSarraV42` procedure in the `bilancarbonsarra.pas` file of the original Pascal code.
Note from
CB : correction of the conversion rate depending on the intensification
level
notes from CB reharding the EvalAssimSarraV42 procedure :
Modif du 04/03/2021 : Prise en compte en plus de la densit� de semis de
l'effet niveau d'intensification NI NI = 1 quand on est � l'optimum du
niveau d'intensification. Dans le cas de situation contr�l� c'est la
fertilit� qui est la clef principale en prenant en r�f�rence la qt� d'azote
(�quivalent phosphore...) optimum Il peut aller � 0 ou �tre sup�rieur � 1 si
situation sur optimum, ie un peu plus de rdt mais � cout trop �lev�... On
�value un nouveau tx de conversion en fn du Ni au travers d'une double
�quation : asympote x gaussienne invers�e Et d'un NI d�fini en fn du
sc�nario de simulation ou des donn�es observ�es. NIYo = D�calage en Y de
l'asymptote NIp = pente de l'asymptote LGauss = Largeur de la Guaussienne
AGauss = Amplitude de la Guaussienne
Conversion qui est la valeur du taux de conversion en situation optimum n'a
plus besoin d'�tre utilis� sinon dans la calibration des param�tres de cette
�quation en absence de donn�es sur ces param�tres on ne met aucune valeur �
NI CF fichier ex IndIntensite_txConv_eq.xls}
Args:
- j (int): An index that represents the current iteration.
- data (dict): A dictionary containing data arrays with the following keys:
- "assimPot" (np.ndarray): An array representing the potential assimilation rate.
- "par" (np.ndarray): An array representing photosynthetically active radiation.
- "lai" (np.ndarray): An array representing the leaf area index.
- "conv" (np.ndarray): An array representing the conversion rate.
- paramVariete (dict): A dictionary containing parameters for the computation of the conversion rate, including:
- "txConversion" (float): The conversion rate.
- "NIYo" (float): The shift in the Y-axis of the asymptote.
- "NIp" (float): The slope of the asymptote.
- "LGauss" (float): The width of the Gaussian curve.
- "AGauss" (float): The amplitude of the Gaussian curve.
- "kdf" (float): The constant used in the computation of `assimPot`.
- paramITK (dict): A dictionary containing the intensification level `NI` (float).
Returns:
- data (dict): The input `data` dictionary with the updated "assimPot" array.
"""
if ~np.isnan(paramITK["NI"]):
#? the following (stupidly long) line was found commented, need to check why and if this is correct
paramVariete["txConversion"] = paramVariete["NIYo"] + paramVariete["NIp"] * (1-np.exp(-paramVariete["NIp"] * paramITK["NI"])) - (np.exp(-0.5*((paramITK["NI"] - paramVariete["LGauss"])/paramVariete["AGauss"])* (paramITK["NI"]- paramVariete["LGauss"])/paramVariete["AGauss"]))/(paramVariete["AGauss"]*2.506628274631)
# NIYo + NIp * (1-exp(-NIp * NI)) - (exp(-0.5*((NI - LGauss)/AGauss)* (NI- LGauss)/AGauss))/(AGauss*2.506628274631)
data["assimPot"][j,:,:] = data["par"][j,:,:] * \
(1-np.exp(-paramVariete["kdf"] * data["lai"][j,:,:])) * \
paramVariete["txConversion"] * 10
else :
data["assimPot"][j,:,:] = data["par"][j,:,:] * \
(1-np.exp(-paramVariete["kdf"] * data["lai"][j,:,:])) * \
data["conv"][j,:,:] * 10
return data
update_bM_and_cM
def update_bM_and_cM(
j,
data,
paramVariete
)
This function returns the updated values of bM and cM.
bM and cM are updated if the delta of aerial biomass is positive, meaning that the plant is gaining aerial biomass, and if the phase is above 1 and below 4 or the phase is below the vegetative phase.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of the bilancarbonsarra.pas files from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_bM_and_cM(j, data, paramVariete):
"""
This function returns the updated values of bM and cM.
bM and cM are updated if the delta of aerial biomass is positive,
meaning that the plant is gaining aerial biomass, and if the phase is
above 1 and below 4 or the phase is below the vegetative phase.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of
the bilancarbonsarra.pas files from the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["bM"][j,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
paramVariete["feuilAeroBase"] - 0.1,
data["bM"][j,:,:],
)
data["cM"][j,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
((paramVariete["feuilAeroPente"] * 1000)/ data["bM"][j,:,:] + 0.78) / 0.75,
data["cM"][j,:,:],
)
return data
update_leaf_biomass
def update_leaf_biomass(
j,
data,
paramVariete
)
For phase above 1 and if the delta of aerial biomass is negative,
meaning that the plant is losing aerial biomass, the leaf biomass is updated as the difference between the leaf biomass and the reallocation minus the delta of aerial biomass multiplied by the reallocation rate in leaves. This value is bound in 0.00000001.
Otherwise, the leaf biomass is not updated.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of the bilancarbonsarra.pas and exmodules 1 & 2.pas files from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_leaf_biomass(j, data, paramVariete):
"""
For phase above 1 and if the delta of aerial biomass is negative,
meaning that the plant is losing aerial biomass, the leaf biomass is
updated as the difference between the leaf biomass and the reallocation
minus the delta of aerial biomass multiplied by the reallocation rate in
leaves. This value is bound in 0.00000001.
Otherwise, the leaf biomass is not updated.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of
the bilancarbonsarra.pas and exmodules 1 & 2.pas files from the original
Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseFeuille"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] < 0),
np.maximum(
0.00000001,
data["biomasseFeuille"][j,:,:] - (data["reallocation"][j,:,:] - data["deltaBiomasseAerienne"][j,:,:]) * paramVariete["pcReallocFeuille"]
),
data["biomasseFeuille"][j,:,:],
)
return data
update_leaf_biomass_all_phases
def update_leaf_biomass_all_phases(
j,
data,
paramVariete
)
summary
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_leaf_biomass_all_phases(j, data, paramVariete):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseFeuille"][j:,:,:] = np.where(
condition_positive_delta_aboveground_biomass_all_phases(j, data),
data["biomasseFeuille"][j,:,:] - data["reallocation"][j,:,:] * paramVariete["pcReallocFeuille"],
data["biomasseFeuille"][j,:,:],
)
return data
update_leaf_biomass_positive_delta_aboveground_biomass
def update_leaf_biomass_positive_delta_aboveground_biomass(
j,
data,
paramVariete
)
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_leaf_biomass_positive_delta_aboveground_biomass(j, data, paramVariete):
"""
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseFeuille"][j:,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
(0.1 + data["bM"][j,:,:] * data["cM"][j,:,:] ** ((data["biomasseAerienne"][j,:,:] - data["rdt"][j,:,:]) / 1000)) \
* (data["biomasseAerienne"][j,:,:] - data["rdt"][j,:,:]),
data["biomasseFeuille"][j,:,:],
)
return data
update_potential_yield
def update_potential_yield(
j,
data,
paramVariete
)
Update the potential yield of the plant.
The potential yield is initialized as an affine function of the delta between the end of the vegetative phase and the end of the flowering stage, plus a linear function of the total biomass at the end of the flowering stage. The potential yield is capped to twice the biomass of the stem to avoid unrealistic values.
The update occurs if the plant is in phase 5 and its phase has changed
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the bilancarbonsarra.pas file from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | An index representing the current time step. | None |
| data | xarray.Dataset | A dataset containing plant data. | None |
| paramVariete | dict | A dictionary containing parameters for the plant variety. | None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The input data with the potential yield updated. |
View Source
def update_potential_yield(j, data, paramVariete):
"""
Update the potential yield of the plant.
The potential yield is initialized as an affine function of the delta
between the end of the vegetative phase and the end of the flowering stage,
plus a linear function of the total biomass at the end of the flowering stage.
The potential yield is capped to twice the biomass of the stem to avoid unrealistic
values.
The update occurs if the plant is in phase 5 and its phase has changed
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the
bilancarbonsarra.pas file from the original Pascal code.
Args:
j (int): An index representing the current time step.
data (xarray.Dataset): A dataset containing plant data.
paramVariete (dict): A dictionary containing parameters for the plant variety.
Returns:
xarray.Dataset: The input `data` with the potential yield updated.
"""
delta_biomass_flowering_ip = data["biomTotStadeFloraison"][j,:,:] - data["biomTotStadeIp"][j,:,:]
data["rdtPot"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5) & (data["changePhase"][j,:,:] == 1),
(paramVariete["KRdtPotA"] * delta_biomass_flowering_ip + paramVariete["KRdtPotB"]) + paramVariete["KRdtBiom"] * data["biomTotStadeFloraison"][j,:,:],
data["rdtPot"][j,:,:],
)
#! phaseDevVeg pas utilisé ? attention c'est un paramètre variétal et pas un jeu de donées
data["rdtPot"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5) & (data["changePhase"][j,:,:] == 1) & (data["rdtPot"][j,:,:] > data["biomasseTige"][j,:,:] * 2) & (paramVariete["phaseDevVeg"] < 6),
data["biomasseTige"][j,:,:] * 2,
data["rdtPot"][j,:,:],
)
return data
update_potential_yield_delta
def update_potential_yield_delta(
j,
data,
paramVariete
)
This function updates the delta potential yield (dRdtPot) of the plant, which is the rate at which the
plant's yield is changing over time. The delta potential yield is calculated as the product of the potential yield, the ratio of actual degree days to maturity, and the ratio of actual transpiration to potential transpiration. The calculation is only done if the plant is in phase 5 and the potential transpiration is above 0. If the potential transpiration is not above 0, the delta potential yield is set to 0. For all other phases, the delta potential yield is unchanged.
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the bilancarbonsarra.pas file from the original Pascal code.
Args: - j (int): an integer index, representing the current step of the simulation - data (xarray dataset): the simulation data, including the current state of the plant - paramVariete (dict): the variety-specific parameters used in the simulation
Returns: - data (xarray dataset): the updated simulation data, including the updated delta potential yield
View Source
def update_potential_yield_delta(j, data, paramVariete):
"""
This function updates the delta potential yield (dRdtPot) of the plant, which is the rate at which the
plant's yield is changing over time. The delta potential yield is calculated as the product of the potential
yield, the ratio of actual degree days to maturity, and the ratio of actual transpiration to potential transpiration.
The calculation is only done if the plant is in phase 5 and the potential transpiration is above 0.
If the potential transpiration is not above 0, the delta potential yield is set to 0.
For all other phases, the delta potential yield is unchanged.
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the
bilancarbonsarra.pas file from the original Pascal code.
Args:
- j (int): an integer index, representing the current step of the simulation
- data (xarray dataset): the simulation data, including the current state of the plant
- paramVariete (dict): the variety-specific parameters used in the simulation
Returns:
- data (xarray dataset): the updated simulation data, including the updated delta potential yield
"""
data["dRdtPot"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5),
np.where(
(data["trPot"][j,:,:] > 0),
np.maximum(
data["rdtPot"][j,:,:] * (data["ddj"][j,:,:] / paramVariete["SDJMatu1"]) * (data["tr"][j,:,:] / data["trPot"][j,:,:]),
data["respMaint"][j,:,:] * 0.15,
),
0,
),
data["dRdtPot"][j,:,:],
)
return data
update_root_biomass
def update_root_biomass(
j,
data
)
Update the root biomass (biomasseRacinaire) for a given time step.
The root biomass is computed as the difference between the total biomass and the aboveground biomass.
This function is based on the EvalBiomasseRacinaire procedure, of the milbilancarbone, exmodules 1 & 2, milbilancarbone.pas file from the original Pascal code
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | Time step index. | None |
| data | xarray.Dataset | Input dataset containing relevant variables. | None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | Updated dataset with the root biomass variable. |
View Source
def update_root_biomass(j, data):
"""
Update the root biomass (biomasseRacinaire) for a given time step.
The root biomass is computed as the difference between the total biomass
and the aboveground biomass.
This function is based on the EvalBiomasseRacinaire procedure, of the
milbilancarbone, exmodules 1 & 2, ***milbilancarbone***.pas file from the
original Pascal code
Args:
j (int): Time step index.
data (xarray.Dataset): Input dataset containing relevant variables.
Returns:
xarray.Dataset: Updated dataset with the root biomass variable.
"""
data["biomasseRacinaire"][j,:,:] = data["biomasseTotale"][j,:,:] - data["biomasseAerienne"][j,:,:]
return data
update_stem_biomass
def update_stem_biomass(
j,
data,
paramVariete
)
For phase above 1 and if the delta of aerial biomass is negative,
meaning that the plant is losing aerial biomass, the stem biomass is updated as the difference between the leaf biomass and the reallocation minus the delta of aerial biomass multiplied by (1-reallocation rate in leaves) (if it's not leaves, it's stems...). This value is bound in 0.00000001.
Otherwise, the stem biomass is not updated.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of the bilancarbonsarra.pas and exmodules 1 & 2.pas files from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_stem_biomass(j, data, paramVariete):
"""
For phase above 1 and if the delta of aerial biomass is negative,
meaning that the plant is losing aerial biomass, the stem biomass is
updated as the difference between the leaf biomass and the reallocation
minus the delta of aerial biomass multiplied by (1-reallocation rate in
leaves) (if it's not leaves, it's stems...). This value is bound in 0.00000001.
Otherwise, the stem biomass is not updated.
This function is adapted from the EvalFeuilleTigeSarrahV4 procedure, of
the bilancarbonsarra.pas and exmodules 1 & 2.pas files from the original
Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
# group 122
data["biomasseTige"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] > 1) & (data["deltaBiomasseAerienne"][j,:,:] < 0),
np.maximum(
0.00000001,
data["biomasseTige"][j,:,:] - (data["reallocation"][j,:,:] - data["deltaBiomasseAerienne"][j,:,:]) * (1 - paramVariete["pcReallocFeuille"]),
),
data["biomasseTige"][j,:,:],
)
return data
update_stem_biomass_all_phases
def update_stem_biomass_all_phases(
j,
data,
paramVariete
)
summary
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_stem_biomass_all_phases(j, data, paramVariete):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseTige"][j:,:,:] = np.where(
condition_positive_delta_aboveground_biomass_all_phases(j, data),
data["biomasseTige"][j,:,:] - (data["reallocation"][j,:,:] * (1- paramVariete["pcReallocFeuille"])),
data["biomasseTige"][j,:,:],
)
return data
update_stem_biomass_positive_delta_aboveground_biomass
def update_stem_biomass_positive_delta_aboveground_biomass(
j,
data,
paramVariete
)
summary
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_stem_biomass_positive_delta_aboveground_biomass(j, data, paramVariete):
"""_summary_
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseTige"][j:,:,:] = np.where(
condition_positive_delta_biomass(j, data, paramVariete),
data["biomasseAerienne"][j,:,:] - data["biomasseFeuille"][j,:,:] - data["rdt"][j,:,:],
data["biomasseTige"][j,:,:],
)
return data
update_total_biomass
def update_total_biomass(
j,
data,
paramVariete,
paramITK
)
Update the Total Biomass of the Plant.
The total biomass is updated based on the plant's current phase and other parameters. If the plant is in phase 2 and there's a change in phase, the total biomass is initialized using crop density, grain yield per plant, and the dry weight of the grain. If the plant is not in phase 2 or there's no change in phase, the total biomass is incremented with the difference between the plant's assimilation and maintenance respiration.
When passing from phase 1 to phase 2, total biomass is initialized. Initialization value is computed from crop density (plants/ha), txResGrain (grain yield per plant), and poidsSecGrain. Otherwise, total biomass is incremented with the difference between plant assimilation assim and maintainance respiration respMaint.
This function is adapted from the EvolBiomTotSarrahV4 procedure, of the bilancarbonsarra.pas file from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | The current time step. | None |
| data | xarray.Dataset | The data for the plant, including variables like "biomasseTotale", "assim", "respMaint", "numPhase", and "changePhase". |
None |
| paramVariete | dict | A dictionary of parameters specific to the plant variety. | None |
| paramITK | dict | A dictionary of inter-tropical convergence zone parameters. | None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The updated data for the plant, including the updated "biomasseTotale" and "deltaBiomasseTotale" variables. |
View Source
def update_total_biomass(j, data, paramVariete, paramITK):
"""
Update the Total Biomass of the Plant.
The total biomass is updated based on the plant's current phase and other parameters.
If the plant is in phase 2 and there's a change in phase, the total biomass is initialized using
crop density, grain yield per plant, and the dry weight of the grain.
If the plant is not in phase 2 or there's no change in phase, the total biomass is incremented with
the difference between the plant's assimilation and maintenance respiration.
When passing from phase 1 to phase 2, total biomass is initialized.
Initialization value is computed from crop density (plants/ha), txResGrain
(grain yield per plant), and poidsSecGrain. Otherwise, total biomass is
incremented with the difference between plant assimilation assim and
maintainance respiration respMaint.
This function is adapted from the EvolBiomTotSarrahV4 procedure, of the
bilancarbonsarra.pas file from the original Pascal code.
Args:
j (int): The current time step.
data (xarray.Dataset): The data for the plant, including variables like
"biomasseTotale", "assim", "respMaint", "numPhase", and "changePhase".
paramVariete (dict): A dictionary of parameters specific to the plant variety.
paramITK (dict): A dictionary of inter-tropical convergence zone parameters.
Returns:
xarray.Dataset: The updated data for the plant, including the updated "biomasseTotale"
and "deltaBiomasseTotale" variables.
"""
data["biomasseTotale"][j:,:,:] = np.where(
(data["numPhase"][j,:,:]==2) & (data["changePhase"][j,:,:]==1),
paramITK["densite"] * np.maximum(1,paramVariete['densOpti']/paramITK['densite']) * paramVariete["txResGrain"] * paramVariete["poidsSecGrain"] / 1000,
data["biomasseTotale"][j,:,:] + (data["assim"][j,:,:] - data["respMaint"][j,:,:]),
)
# we may want to drop this variable and use the raw computation instead
data["deltaBiomasseTotale"][j:,:,:] = (data["assim"][j,:,:] - data["respMaint"][j,:,:])
return data
update_total_biomass_at_flowering_stage
def update_total_biomass_at_flowering_stage(
j,
data
)
This function updates the total biomass of the plant at the end of the
flowering stage (biomTotStadeFloraison).
If the plant is in phase 5, and the phase has changed, then the total biomass is copied to the biomTotStadeFloraison variable.
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the bilancarbonsarra.pas file from the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_total_biomass_at_flowering_stage(j, data):
"""
This function updates the total biomass of the plant at the end of the
flowering stage (biomTotStadeFloraison).
If the plant is in phase 5, and the phase has changed, then the total
biomass is copied to the biomTotStadeFloraison variable.
This function is adapted from the EvalRdtPotRespSarV42 procedure, of
the bilancarbonsarra.pas file from the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomTotStadeFloraison"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5) & (data["changePhase"][j,:,:] == 1),
data["biomasseTotale"][j,:,:],
data["biomTotStadeFloraison"][j,:,:],
)
return data
update_total_biomass_stade_ip
def update_total_biomass_stade_ip(
j,
data
)
Update the total biomass of the plant at the end of the vegetative phase (ip = "initiation paniculaire").
If the plant has reached phase 4 and has just changed phase, the current total biomass will be copied to the "biomTotStadeIp" variable, which represents the total biomass at the end of the vegetative phase (initiation paniculaire).
This function is adapted from the EvalRdtPotRespSarV42 procedure, of the bilancarbonsarra.pas file from the original Pascal code.
Args: j (int): Timestep index. data (xarray.Dataset): Input dataset.
Returns: xarray.Dataset: The updated dataset with the "biomTotStadeIp" variable updated.
View Source
def update_total_biomass_stade_ip(j, data):
"""
Update the total biomass of the plant at the end of the vegetative phase (ip = "initiation paniculaire").
If the plant has reached phase 4 and has just changed phase, the current
total biomass will be copied to the "biomTotStadeIp" variable, which represents
the total biomass at the end of the vegetative phase (initiation paniculaire).
This function is adapted from the EvalRdtPotRespSarV42 procedure, of
the bilancarbonsarra.pas file from the original Pascal code.
Args:
j (int): Timestep index.
data (xarray.Dataset): Input dataset.
Returns:
xarray.Dataset: The updated dataset with the "biomTotStadeIp" variable updated.
"""
data["biomTotStadeIp"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 4) & (data["changePhase"][j,:,:] == 1),
data["biomasseTotale"][j,:,:],
data["biomTotStadeIp"][j,:,:],
)
return data
update_vegetative_biomass
def update_vegetative_biomass(
j,
data
)
summary
This function is adapted from the EvalBiomasseVegetati procedure from the copie milbilancarbon, exmodules 1 & 2, milbilancarbone file of the original Pascal code.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | type | description | None |
| data | type | description | None |
Returns:
| Type | Description |
|---|---|
| type | description |
View Source
def update_vegetative_biomass(j, data):
"""_summary_
This function is adapted from the EvalBiomasseVegetati procedure from the copie milbilancarbon, exmodules 1 & 2, ***milbilancarbone*** file
of the original Pascal code.
Args:
j (_type_): _description_
data (_type_): _description_
Returns:
_type_: _description_
"""
data["biomasseVegetative"][j:,:,:] = (data["biomasseTige"][j,:,:] + data["biomasseFeuille"][j,:,:])
return data
update_yield_during_filling_phase
def update_yield_during_filling_phase(
j,
data
)
This function updates the yield value during the filling phase.
During the filling phase (numPhase == 5), the yield is updated by
incrementing it with the sum of deltaBiomasseAerienne and reallocation,
bounded by 0 and dRdtPot (daily potential yield). The construction of yield
is done during phase 5 only, from the variation of aerial biomass and
reallocation, with a maximum of dRdtPot.
This function is adapted from the EvolDayRdtSarraV3 procedure from the bilancarbonesarra, exmodules 1 & 2.pas file of the original Pascal code.
Notes : On tend vers le potentiel en fn du rapport des degresJours/sumDegresJours pour la phase de remplissage. Frein sup fn du flux de sève estimé par le rapport Tr/TrPot. dRdtPot = RdtPotDuJour
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
| j | int | The time step at which the calculation starts. | None |
| data | xarray.Dataset | The data that contains all variables. | None |
Returns:
| Type | Description |
|---|---|
| xarray.Dataset | The input data with updated yield values. |
View Source
def update_yield_during_filling_phase(j, data):
"""
This function updates the yield value during the filling phase.
During the filling phase (numPhase == 5), the yield is updated by
incrementing it with the sum of `deltaBiomasseAerienne` and `reallocation`,
bounded by 0 and `dRdtPot` (daily potential yield). The construction of yield
is done during phase 5 only, from the variation of aerial biomass and
reallocation, with a maximum of `dRdtPot`.
This function is adapted from the EvolDayRdtSarraV3 procedure from the
***bilancarbonesarra***, exmodules 1 & 2.pas file of the original Pascal
code.
Notes :
On tend vers le potentiel en fn du rapport des degresJours/sumDegresJours
pour la phase de remplissage. Frein sup fn du flux de sève estimé par le
rapport Tr/TrPot.
dRdtPot = RdtPotDuJour
Args:
j (int): The time step at which the calculation starts.
data (xarray.Dataset): The data that contains all variables.
Returns:
xarray.Dataset: The input data with updated yield values.
"""
data["rdt"][j:,:,:] = np.where(
(data["numPhase"][j,:,:] == 5),
data["rdt"][j,:,:] + np.minimum(data["dRdtPot"][j,:,:], np.maximum(0.0, data["deltaBiomasseAerienne"][j,:,:]) + data['reallocation'][j,:,:]),
data["rdt"][j,:,:],
)
return data
variable_dict
def variable_dict(
)
Retrieve the dictionary of variables in the dataset with their respective units.
Returns:
| Type | Description |
|---|---|
| dict | A dictionary containing the variables and their units, where the keys are the variable names and the values are the respective units. |
View Source
def variable_dict():
"""
Retrieve the dictionary of variables in the dataset with their respective units.
Returns:
dict: A dictionary containing the variables and their units, where the keys are the variable names and the values are the respective units.
"""
variables = {
# climate
"ddj": ["daily thermal time", "°C.j"],
"sdj": ["sum of thermal time since beginning of emergence", "°C.j"],
# phenology
"changePhase": ["indicator of phase transition day", "binary"],
"numPhase": ["number of phenological stage", "arbitrary units"],
"initPhase": ["indicator of performed phase transition", "binary"],
"phasePhotoper": ["photoperiodic phase indicator", "binary"],
"seuilTempPhaseSuivante": ["sum of thermal time needed to reach the next phenological phase", "°C.j"],
"sommeDegresJourPhasePrec": ["sum of thermal time needed to reach the previous phenological phase", "°C.j"],
"seuilTempPhasePrec": ["sum of thermal time needed to reach the previous phenological phase", "°C.j"],
# carbon balance
"assim": ["plant biomass assimilation", "kg/ha"],
"assimPot": ["plant potential biomass assimilation", "kg/ha"],
"bM": ["net growth rate of living biomass", "kg/(m².d)"],
"cM": ["net growth rate of dead biomass", "kg/(m².d)"],
"rdt": ["grain yield", "kg/ha"],
"rdtPot": ["potential grain yield", "kg/ha"],
"reallocation": ["amount of assimilates reallocated to the yield (supply < demand)", "kg/ha"],
"respMaint": ["amount of assimilates consumed by maintainance respiration", "kg/ha"],
"manqueAssim": ["deficit in assimilates (demand - supply)", "kg/ha"],
# biomass
"biomTotStadeFloraison": ["total biomass of the plant at the end of the flowering stage", "kg/ha"],
"biomTotStadeIp": ["total biomass at the panicle initiation stage", "kg/ha"],
"deltaBiomasseAerienne": ["increment of aerial biomass in one day", "kg/(ha.d)"],
"deltaBiomasseFeuilles": ["increment of leaf biomass in one day", "kg/(ha.d)"],
"biomasseAerienne": ["total aerial biomass", "kg/ha"],
"biomasseVegetative": ["total vegetative biomass", "kg/ha"],
"biomasseTotale": ["total biomass", "kg/ha"],
"biomasseTige": ["total stem biomass", "kg/ha"],
"biomasseRacinaire": ["total root biomass", "kg/ha"],
"biomasseFeuille": ["total leaf biomass", "kg/ha"],
"deltaBiomasseTotale": ["increment of total biomass in one day", "kg/(ha.d)"],
# evapotranspiration
"kce": ["fraction of kc attributable to soil evaporation","decimal percentage"],
"kcp": ["fraction of kc attributable to plant transpiration","decimal percentage"],
"kcTot": ["total crop coefficient",""],
"tr": ["actual crop transpiration","mm/d"],
"trPot": ["potential crop transpiration","mm/d"],
"trSurf": ["",""],
# water balance
"consoRur": ["consumption of water stored in the root system", "mm"],
"water_captured_by_mulch" : ["water captured by the mulch in one day","mm"],
"available_water" : ["available water, sum of rainfall and total irrigation for the day","mm"],
"eauTranspi": ["water available for transpiration from the surface reservoir","mm"],
"correctedIrrigation" : ["corrected irrigation amount","mm/d"],
"cstr" : ["drought stress coefficient", "arbitrary unit"],
"dayVrac" : ["modulated daily root growth","mm/day"],
"delta_root_tank_capacity": ["change in root system water reserve","mm"],
"drainage": ["drainage","mm"],
#// "etm": ["evapotranspiration from the soil moisture","mm/d"],
"etp": ["potential evapotranspiration from the soil moisture","mm/d"],
#// "etr": ["reference evapotranspiration","mm/d"],
"evap": ["evaporation from the soil moisture","mm/d"],
"evapPot": ["potential evaporation from the soil moisture","mm/d"],
"FEMcW": ["water fraction in soil volume explored by the root system","none"],
"fesw": ["fraction of available surface water","decimal percentage"],
"irrigTotDay" : ["total irrigation for the day","mm"],
"vRac" : ["reference daily root growth","mm/day"],
"ftsw": ["fraction of transpirable surface water","decimal percentage"],
"runoff" : ["daily water runoff","mm/d"],
"pFact": ["FAO reference for critical FTSW value for transpiration response","none"],
# water tanks
"irrigation_tank_stock" : ["current stock of water in the irrigation tank","mm"], #! renaming stockIrr to irrigation_tank_stock
"mulch_water_stock" : ["water stored in crop residues (mulch)","mm"], #! renaming stockMc to mulch_water_stock
"root_tank_stock": ["current stock of water in the root system tank","mm"], #! renaming stRu to root_tank_stock
"total_tank_capacity": ["total capacity of the root system tank","mm"], #! renaming stRuMax to total_tank_capacity
"stRur": ["",""], # ["previous season's root system tank stock","mm"],
"previous_root_tank_capacity": ["previous season's root system tank capacity","mm"], #! renaming stRurMaxPrec to previous_root_tank_capacity
"previous_root_tank_stock": ["previous day's root system tank stock","mm"],
"stRurSurf": ["surface root system tank stock","mm"],
"surface_tank_stock": ["current stock of water in the surface root system tank","mm"], #! renaming stRuSurf to surface_tank_stock
"stRuSurfPrec": ["previous day's surface root system tank stock","mm"],
"delta_total_tank_stock": ["change in the total root system tank stock","mm"], #! renaming stRuVar to delta_total_tank_stock
"irrigation_tank_capacity" : ["irrigation tank capacity","mm"], #! renaming ruIrr to irrigation_tank_capacity
"ruRac": ["Water column that can potentially be strored in soil volume explored by root system","mm"],
"conv": ["",""],
"KAssim": ["",""],
"dayBiomLeaf": ["daily growth of leaf biomass","kg/ha/d"],
"dRdtPot": ["daily potential demand from yield","kg/ha/d"],
"FeuilleUp": ["",""],
"kRespMaint": ["",""],
"LitFeuille": ["",""],
"nbJourCompte": ["",""],
"nbjStress": ["",""],
"NbUBT": ["",""],
"sla": ["",""],
"stockRac": ["",""],
"sumPP": ["",""],
"TigeUp": ["",""],
"UBTCulture": ["",""],
"lai":["leaf area index","m2/m2"],
# experimental
"Ncrit": ["",""],
}
return variables